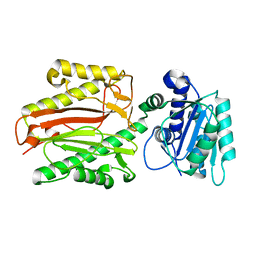
5E5B
 
 | |
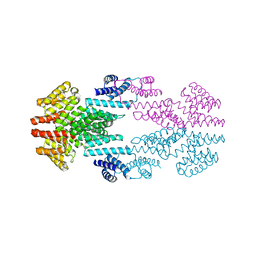
6T3H
 
 | |
7Y9H
 
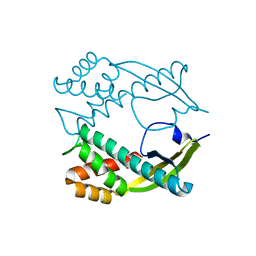
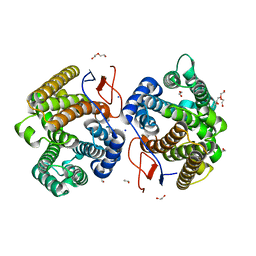 | | Crystal structure of diterpene synthase VenA from Streptomyces venezuelae ATCC 15439 | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Diterpene synthase VenA, ... | | Authors: | Zhang, L.L, Xie, Z.Z, Huang, J.-W, Hu, Y.C, Liu, W.D, Yang, Y, Chen, C.-C, Guo, R.-T. | | Deposit date: | 2022-06-24 | | Release date: | 2023-06-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Molecular insights into the catalytic promiscuity of a bacterial diterpene synthase.
Nat Commun, 14, 2023
|
|
3GUQ
 
 | | Crystal structure of novel carcinogenic factor of H. pylori | | Descriptor: | Putative uncharacterized protein | | Authors: | Tsurumura, T, Tsuge, H, Utsunomiya, H, Kise, D, Kuzuhara, T, Fujiki, H, Suganuma, M. | | Deposit date: | 2009-03-30 | | Release date: | 2009-09-22 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Structural basis for the Helicobacter pylori-carcinogenic TNF-alpha-inducing protein.
Biochem.Biophys.Res.Commun., 388, 2009
|
|
8QWK
 
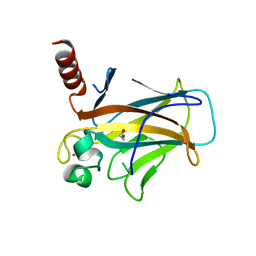 | | Structure of p53 cancer mutant Y126C | | Descriptor: | 1,2-ETHANEDIOL, Cellular tumor antigen p53, ZINC ION | | Authors: | Markl, A.M, Balourdas, D.I, Kraemer, A, Knapp, S, Joerger, A.C, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-10-19 | | Release date: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Structural basis of p53 inactivation by cavity-creating cancer mutations and its implications for the development of mutant p53 reactivators.
Cell Death Dis, 15, 2024
|
|
8QWP
 
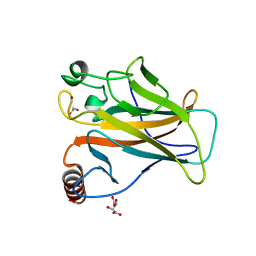 | | Structure of p53 cancer mutant Y236C | | Descriptor: | 1,2-ETHANEDIOL, Cellular tumor antigen p53, L(+)-TARTARIC ACID, ... | | Authors: | Balourdas, D.I, Knapp, S, Joerger, A.C, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-10-19 | | Release date: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis of p53 inactivation by cavity-creating cancer mutations and its implications for the development of mutant p53 reactivators.
Cell Death Dis, 15, 2024
|
|
3GSI
 
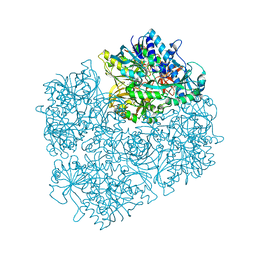 | | Crystal structure of D552A dimethylglycine oxidase mutant of Arthrobacter globiformis in complex with tetrahydrofolate | | Descriptor: | (6S)-5,6,7,8-TETRAHYDROFOLATE, FLAVIN-ADENINE DINUCLEOTIDE, MAGNESIUM ION, ... | | Authors: | Tralau, T, Lafite, P, Levy, C, Combe, J.P, Scrutton, N.S, Leys, D. | | Deposit date: | 2009-03-27 | | Release date: | 2009-04-14 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | An internal reaction chamber in dimethylglycine oxidase provides efficient protection from exposure to toxic formaldehyde.
J.Biol.Chem., 284, 2009
|
|
7NH4
 
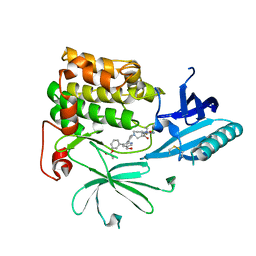 | |
7MQD
 
 | | Bartonella henselae NrnC complexed with pAGG. D4 symmetry. | | Descriptor: | NanoRNase C, RNA (5'-R(P*AP*GP*G)-3') | | Authors: | Lormand, J.D, Brownfield, B, Fromme, J.C, Sondermann, H. | | Deposit date: | 2021-05-05 | | Release date: | 2021-09-15 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (3.25 Å) | | Cite: | Structural characterization of NrnC identifies unifying features of dinucleotidases.
Elife, 10, 2021
|
|
8QWO
 
 | | Structure of p53 cancer mutant Y234C | | Descriptor: | Cellular tumor antigen p53, GLYCEROL, ZINC ION | | Authors: | Balourdas, D.I, Knapp, S, Joerger, A.C, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-10-19 | | Release date: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Structural basis of p53 inactivation by cavity-creating cancer mutations and its implications for the development of mutant p53 reactivators.
Cell Death Dis, 15, 2024
|
|
8ZVF
 
 | | AtALMT9 plus high malate in low pH | | Descriptor: | 1,2-DILAUROYL-SN-GLYCERO-3-PHOSPHATE, Aluminum-activated malate transporter 9 | | Authors: | Gong, D.S. | | Deposit date: | 2024-06-11 | | Release date: | 2024-09-25 | | Method: | ELECTRON MICROSCOPY (3.59 Å) | | Cite: | Structural insight into the Arabidopsis vacuolar anion channel ALMT9 shows clade specificity.
Cell Rep, 43, 2024
|
|
7MQC
 
 | | Bartonella henselae NrnC bound to pGG. C1 reconstruction. | | Descriptor: | NanoRNase C, RNA (5'-R(P*GP*G)-3') | | Authors: | Lormand, J.D, Brownfield, B, Fromme, J.C, Sondermann, H. | | Deposit date: | 2021-05-05 | | Release date: | 2021-09-15 | | Last modified: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (3.64 Å) | | Cite: | Structural characterization of NrnC identifies unifying features of dinucleotidases.
Elife, 10, 2021
|
|
7MPM
 
 | | Bartonella henselae NrnC bound to pAA | | Descriptor: | 5'-phosphorylated AA, NanoRNase C | | Authors: | Lormand, J.D, Sondermann, H. | | Deposit date: | 2021-05-04 | | Release date: | 2021-09-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural characterization of NrnC identifies unifying features of dinucleotidases.
Elife, 10, 2021
|
|
5NK7
 
 | | Crystal Structure of Ephrin A2 (EphA2) Receptor Protein Kinase with Compound 2a | | Descriptor: | Ephrin type-A receptor 2, ~{N}-(2-chloranyl-6-methyl-phenyl)-2-[[3-[[(3~{R},4~{R})-3-fluoranylpiperidin-4-yl]carbamoyl]phenyl]amino]-1,3-thiazole-5-carboxamide | | Authors: | Kudlinzki, D, Linhard, V.L, Witt, K, Gande, S.L, Saxena, K, Heinzlmeir, S, Medard, G, Kuester, B, Schwalbe, H. | | Deposit date: | 2017-03-31 | | Release date: | 2017-06-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.889 Å) | | Cite: | Chemoproteomics-Aided Medicinal Chemistry for the Discovery of EPHA2 Inhibitors.
ChemMedChem, 12, 2017
|
|
5NKF
 
 | | Crystal Structure of Ephrin A2 (EphA2) Receptor Protein Kinase with Compound 3b | | Descriptor: | Ephrin type-A receptor 2, ~{N}-(2-chloranyl-6-methyl-phenyl)-2-[[3-(piperidin-4-ylcarbamoyl)-5-(trifluoromethyl)phenyl]amino]-1,3-thiazole-5-carboxamide | | Authors: | Kudlinzki, D, Linhard, V.L, Witt, K, Gande, S.L, Saxena, K, Heinzlmeir, S, Medard, G, Kuester, B, Schwalbe, H. | | Deposit date: | 2017-03-31 | | Release date: | 2017-06-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.099 Å) | | Cite: | Chemoproteomics-Aided Medicinal Chemistry for the Discovery of EPHA2 Inhibitors.
ChemMedChem, 12, 2017
|
|
7MPN
 
 | | Bartonella henselae NrnC bound to pGC | | Descriptor: | 5'-phosphorylated GC, NanoRNase C | | Authors: | Lormand, J.D, Sondermann, H. | | Deposit date: | 2021-05-04 | | Release date: | 2021-09-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Structural characterization of NrnC identifies unifying features of dinucleotidases.
Elife, 10, 2021
|
|
5DX5
 
 | | Crystal structure of methionine gamma-lyase from Clostridium sporogenes | | Descriptor: | CHLORIDE ION, Methionine gamma-lyase, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Revtovich, S.V, Nikulin, A.D, Morozova, E.A, Anufrieva, N.V, Demidkina, T.V. | | Deposit date: | 2015-09-23 | | Release date: | 2016-01-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Structure of methionine gamma-lyase from Clostridium sporogenes.
Acta Crystallogr.,Sect.F, 72, 2016
|
|
5NK9
 
 | | Crystal Structure of Ephrin A2 (EphA2) Receptor Protein Kinase with Compound 2e | | Descriptor: | (2~{Z})-~{N}-(2-chloranyl-6-methyl-phenyl)-2-[3-[(4-methyl-4-oxidanyl-cyclohexyl)carbamoyl]phenyl]imino-1,3-thiazolidine-5-carboxamide, Ephrin type-A receptor 2 | | Authors: | Kudlinzki, D, Linhard, V.L, Witt, K, Gande, S.L, Saxena, K, Heinzlmeir, S, Medard, G, Kuester, B, Schwalbe, H. | | Deposit date: | 2017-03-31 | | Release date: | 2017-06-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.588 Å) | | Cite: | Chemoproteomics-Aided Medicinal Chemistry for the Discovery of EPHA2 Inhibitors.
ChemMedChem, 12, 2017
|
|
1T3J
 
 | | Mitofusin domain HR2 V686M/I708M mutant | | Descriptor: | mitofusin 1 | | Authors: | Koshiba, T, Detmer, S.A, Kaiser, J.T, Chen, H, McCaffery, J.M, Chan, D.C. | | Deposit date: | 2004-04-26 | | Release date: | 2004-08-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis of mitochondrial tethering by mitofusin complexes
Science, 305, 2004
|
|
5EB6
 
 | |
3CE7
 
 | | Crystal structure of toxoplasma specific mitochondrial acyl carrier protein, 59.m03510 | | Descriptor: | Specific mitochondrial acyl carrier protein | | Authors: | Wernimont, A.K, Dong, A, Yang, C, Khuu, C, Lam, A, Brand, V, Kozieradzki, I, Cossar, D, Arrowsmith, C.H, Bountra, C, Edwards, A.M, Weigelt, J, Bochkarev, A, Hui, R, Qiu, W, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-02-28 | | Release date: | 2008-03-25 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Crystal structure of toxoplasma specific mitochondrial acyl carrier protein, 59.m03510.
To be Published
|
|
5NKI
 
 | | Crystal Structure of Ephrin A2 (EphA2) Receptor Protein Kinase with Compound 4b | | Descriptor: | Ephrin type-A receptor 2, ~{N}-(2-chloranyl-6-methyl-phenyl)-2-[(3-methylsulfonyl-5-morpholin-4-yl-phenyl)amino]-1,3-thiazole-5-carboxamide | | Authors: | Kudlinzki, D, Linhard, V.L, Witt, K, Gande, S.L, Saxena, K, Heinzlmeir, S, Medard, G, Kuester, B, Schwalbe, H. | | Deposit date: | 2017-03-31 | | Release date: | 2017-06-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.675 Å) | | Cite: | Chemoproteomics-Aided Medicinal Chemistry for the Discovery of EPHA2 Inhibitors.
ChemMedChem, 12, 2017
|
|
8QWN
 
 | | Structure of p53 cancer mutant Y220C (hexagonal crystal form) | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Cellular tumor antigen p53, ... | | Authors: | Balourdas, D.I, Knapp, S, Joerger, A.C, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-10-19 | | Release date: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Structural basis of p53 inactivation by cavity-creating cancer mutations and its implications for the development of mutant p53 reactivators.
Cell Death Dis, 15, 2024
|
|
8QWM
 
 | | Structure of p53 cancer mutant Y205C | | Descriptor: | 1,2-ETHANEDIOL, Cellular tumor antigen p53, L(+)-TARTARIC ACID, ... | | Authors: | Balourdas, D.I, Knapp, S, Joerger, A.C, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-10-19 | | Release date: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Structural basis of p53 inactivation by cavity-creating cancer mutations and its implications for the development of mutant p53 reactivators.
Cell Death Dis, 15, 2024
|
|
7SEZ
 
 | | Crystal structure of Vaccinia Virus decapping enzyme D9 in complex with m7GDP | | Descriptor: | 7N-METHYL-8-HYDROGUANOSINE-5'-DIPHOSPHATE, DNA repair NTP-phosphohydrolase, SODIUM ION | | Authors: | Peters, J.K, Tibble, R.W, Warminski, M, Jemielity, J, Gross, J.D. | | Deposit date: | 2021-10-02 | | Release date: | 2022-03-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.70001245 Å) | | Cite: | Structure of the poxvirus decapping enzyme D9 reveals its mechanism of cap recognition and catalysis.
Structure, 30, 2022
|
|
