5MLX
 
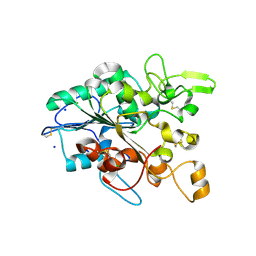 | | Open loop conformation of PhaZ7 Y105E mutant | | Descriptor: | CHLORIDE ION, PHB depolymerase PhaZ7, SODIUM ION | | Authors: | Kellici, T, Mavromoustakos, T, Jendrossek, D, Papageorgiou, A.C. | | Deposit date: | 2016-12-08 | | Release date: | 2017-05-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure analysis, covalent docking, and molecular dynamics calculations reveal a conformational switch in PhaZ7 PHB depolymerase.
Proteins, 85, 2017
|
|
4X0O
 
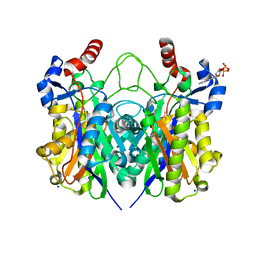 | | Beta-ketoacyl-(acyl carrier protein) synthase III-2 (FabH2) from Vibrio cholerae soaked with Acetyl-CoA | | Descriptor: | 3-oxoacyl-[acyl-carrier-protein] synthase 3 protein 2, COENZYME A, MALONATE ION, ... | | Authors: | Hou, J, Chruszcz, M, Zheng, H, Cooper, D.R, Chordia, M.D, Zimmerman, M.D, Anderson, W.F, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-11-21 | | Release date: | 2014-12-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and enzymatic studies of beta-ketoacyl-(acyl carrier protein) synthase III (FabH) from Vibrio cholerae
to be published
|
|
7P03
 
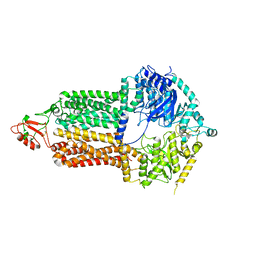 | | Cryo-EM structure of Pdr5 from Saccharomyces cerevisiae in inward-facing conformation without nucleotides | | Descriptor: | Pleiotropic ABC efflux transporter of multiple drugs | | Authors: | Szewczak-Harris, A, Wagner, M, Du, D, Schmitt, L, Luisi, B.F. | | Deposit date: | 2021-06-29 | | Release date: | 2021-11-10 | | Method: | ELECTRON MICROSCOPY (3.45 Å) | | Cite: | Structure and efflux mechanism of the yeast pleiotropic drug resistance transporter Pdr5.
Nat Commun, 12, 2021
|
|
7TR9
 
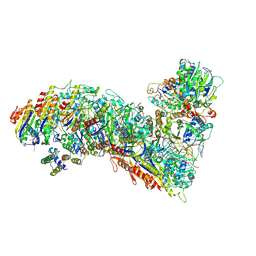 | | Cascade complex from type I-A CRISPR-Cas system | | Descriptor: | CRISPR-associated endonuclease Cas3-HD, CRISPR-associated helicase Cas3, Cas11a, ... | | Authors: | Hu, C, Ni, D, Nam, K.H, Majumdar, S, McLean, J, Stahlberg, H, Terns, M, Ke, A. | | Deposit date: | 2022-01-28 | | Release date: | 2022-08-10 | | Last modified: | 2022-08-24 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Allosteric control of type I-A CRISPR-Cas3 complexes and establishment as effective nucleic acid detection and human genome editing tools.
Mol.Cell, 82, 2022
|
|
6NSS
 
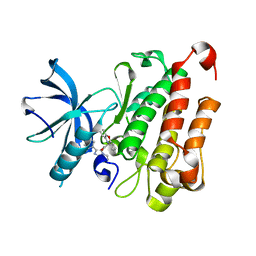 | | TRK-A IN COMPLEX WITH LIGAND 6 | | Descriptor: | High affinity nerve growth factor receptor, N-(8-methyl-2-phenylimidazo[1,2-a]pyrazin-3-yl)-2-(10H-phenoxazin-10-yl)acetamide | | Authors: | Subramanian, G, Brown, D.G. | | Deposit date: | 2019-01-25 | | Release date: | 2019-05-22 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Deciphering the Allosteric Binding Mechanism of the Human Tropomyosin Receptor Kinase A ( hTrkA) Inhibitors.
Acs Chem.Biol., 14, 2019
|
|
7JWY
 
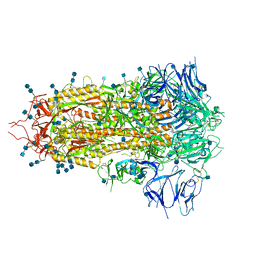 | | Structure of SARS-CoV-2 spike at pH 4.5 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Zhou, T, Tsybovsky, Y, Kwong, P.D. | | Deposit date: | 2020-08-26 | | Release date: | 2020-11-25 | | Last modified: | 2021-12-15 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Cryo-EM Structures of SARS-CoV-2 Spike without and with ACE2 Reveal a pH-Dependent Switch to Mediate Endosomal Positioning of Receptor-Binding Domains.
Cell Host Microbe, 28, 2020
|
|
7P04
 
 | | Cryo-EM structure of Pdr5 from Saccharomyces cerevisiae in inward-facing conformation with ADP/ATP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Pleiotropic ABC efflux transporter of multiple drugs | | Authors: | Szewczak-Harris, A, Wagner, M, Du, D, Schmitt, L, Luisi, B.F. | | Deposit date: | 2021-06-29 | | Release date: | 2021-11-10 | | Method: | ELECTRON MICROSCOPY (2.85 Å) | | Cite: | Structure and efflux mechanism of the yeast pleiotropic drug resistance transporter Pdr5.
Nat Commun, 12, 2021
|
|
6AQO
 
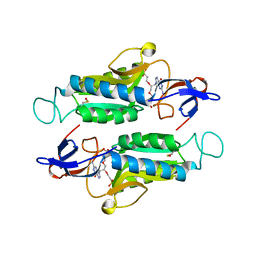 | | Crystal structure of hypoxanthine-guanine-xanthine phosphorybosyltranferase in complex with {[(2S)-3-(2-amino-6-oxo-1,6-dihydro-9H-purin-9-yl)propane-1,2-diyl]bis(oxyethane-2,1-diyl)}bis(phosphonic acid) | | Descriptor: | Hypoxanthine-guanine phosphoribosyltransferase, MAGNESIUM ION, {[(2S)-3-(2-amino-6-oxo-1,6-dihydro-9H-purin-9-yl)propane-1,2-diyl]bis(oxyethane-2,1-diyl)}bis(phosphonic acid) | | Authors: | Teran, D, Gudday, L.W. | | Deposit date: | 2017-08-21 | | Release date: | 2018-03-14 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Evaluation of the Trypanosoma brucei 6-oxopurine salvage pathway as a potential target for drug discovery.
PLoS Negl Trop Dis, 12, 2018
|
|
6NB8
 
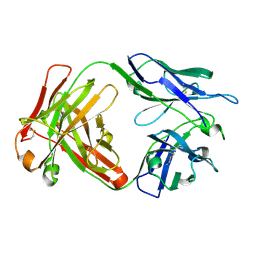 | | Crystal structure of anti- SARS-CoV human neutralizing S230 antibody Fab fragment | | Descriptor: | S230 antigen-binding (Fab) fragment, heavy chain, light chain | | Authors: | Walls, A.J, Xiong, X, Park, Y.J, Tortorici, M.A, Snijder, J, Quispe, J, Cameroni, E, Gopal, R, Dai, M, Lanzavecchia, A, Zambon, M, Rey, F.A, Corti, D, Veesler, D, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2018-12-06 | | Release date: | 2019-02-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Unexpected Receptor Functional Mimicry Elucidates Activation of Coronavirus Fusion.
Cell, 176, 2019
|
|
5F96
 
 | |
8DFS
 
 | | type I-C Cascade bound to AcrIF2 | | Descriptor: | Anti-CRISPR protein 30, CRISPR-associated protein, CT1133 family, ... | | Authors: | O'Brien, R.E, Bravo, J.P.K, Ramos, D, Hibshman, G.N, Wright, J.T, Taylor, D.W. | | Deposit date: | 2022-06-22 | | Release date: | 2023-02-22 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural snapshots of R-loop formation by a type I-C CRISPR Cascade.
Mol.Cell, 83, 2023
|
|
6TIC
 
 | |
6TIN
 
 | |
5NMH
 
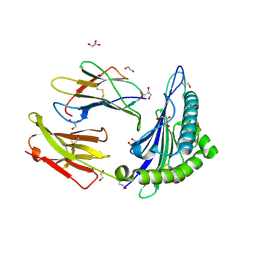 | | HLA A02 presenting SLYNTIATL | | Descriptor: | 1,2-ETHANEDIOL, Beta-2-microglobulin, GLYCEROL, ... | | Authors: | Rizkallah, P.J, Cole, D.K, Fuller, A, Sewell, A.K. | | Deposit date: | 2017-04-05 | | Release date: | 2017-11-15 | | Last modified: | 2017-12-20 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Dual Molecular Mechanisms Govern Escape at Immunodominant HLA A2-Restricted HIV Epitope.
Front Immunol, 8, 2017
|
|
8DFO
 
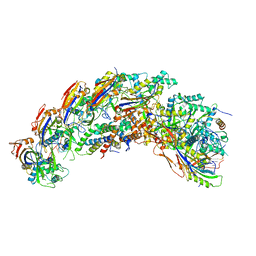 | | type I-C Cascade bound to AcrIC4 | | Descriptor: | AcrIC4, CRISPR-associated protein, CT1133 family, ... | | Authors: | O'Brien, R.E, Bravo, J.P.K, Ramos, D, Hibshman, G.N, Wright, J.T, Taylor, D.W. | | Deposit date: | 2022-06-22 | | Release date: | 2023-02-15 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural snapshots of R-loop formation by a type I-C CRISPR Cascade.
Mol.Cell, 83, 2023
|
|
6ZTY
 
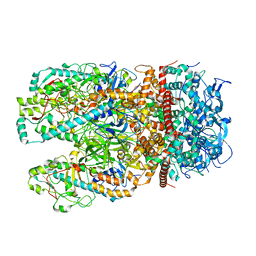 | |
6WCX
 
 | |
8UM6
 
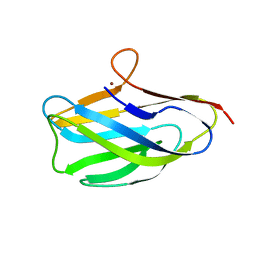 | |
5NMW
 
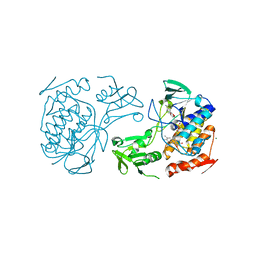 | | Crystal Structure of the pyrrolizidine alkaloid N-oxygenase from Zonocerus variegatus in complex with FAD | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Flavin-containing monooxygenase, MAGNESIUM ION | | Authors: | Scheidig, A, Kubitza, C, Faust, A, Ober, D. | | Deposit date: | 2017-04-07 | | Release date: | 2018-03-14 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Crystal structure of pyrrolizidine alkaloid N-oxygenase from the grasshopper Zonocerus variegatus.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
8DFA
 
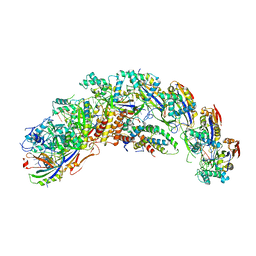 | | type I-C Cascade bound to ssDNA target | | Descriptor: | CRISPR-associated protein, CT1133 family, TM1801 family, ... | | Authors: | O'Brien, R.E, Bravo, J.P.K, Ramos, D, Hibshman, G.N, Wright, J.T, Taylor, D.W. | | Deposit date: | 2022-06-21 | | Release date: | 2023-02-22 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural snapshots of R-loop formation by a type I-C CRISPR Cascade.
Mol.Cell, 83, 2023
|
|
8A4V
 
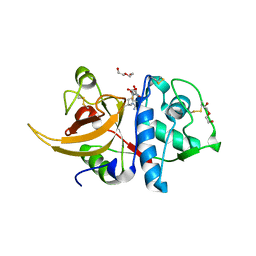 | | Crystal structure of human cathepsin L with covalently bound E-64 | | Descriptor: | Cathepsin L, DI(HYDROXYETHYL)ETHER, DIMETHYL SULFOXIDE, ... | | Authors: | Falke, S, Lieske, J, Guenther, S, Reinke, P.Y.A, Ewert, W, Loboda, J, Karnicar, K, Usenik, A, Lindic, N, Sekirnik, A, Chapman, H.N, Hinrichs, W, Turk, D, Meents, A. | | Deposit date: | 2022-06-13 | | Release date: | 2023-07-05 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural Elucidation and Antiviral Activity of Covalent Cathepsin L Inhibitors.
J.Med.Chem., 67, 2024
|
|
6TIH
 
 | |
7JV6
 
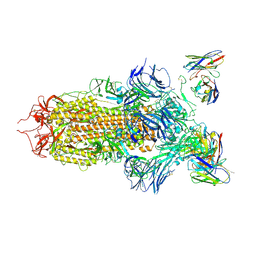 | | SARS-CoV-2 spike in complex with the S2H13 neutralizing antibody (closed conformation) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, S2H13 Fab heavy chain, ... | | Authors: | Park, Y.J, Tortorici, M.A, Walls, A.C, Czudnochowski, N, Seattle Structural Genomics Center for Infectious Disease (SSGCID), Snell, G, Veesler, D. | | Deposit date: | 2020-08-20 | | Release date: | 2020-10-14 | | Last modified: | 2021-06-23 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Mapping Neutralizing and Immunodominant Sites on the SARS-CoV-2 Spike Receptor-Binding Domain by Structure-Guided High-Resolution Serology.
Cell, 183, 2020
|
|
6FWL
 
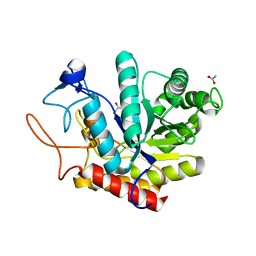 | | Structure of an E333Q variant of the GH99 endo-alpha-mannanase from Bacteroides xylanisolvens in complex alpha-Glc-1,3-(1,2-anhydro-carba-mannose) | | Descriptor: | (1~{S},2~{S},3~{R},4~{R},6~{R})-4-(hydroxymethyl)-7-oxabicyclo[4.1.0]heptane-2,3-diol, ACETATE ION, Glycosyl hydrolase family 71, ... | | Authors: | Sobala, L.F, Speciale, G, Hakki, Z, Fernandes, P.Z, Raich, L, Rojas-Cervellera, V, Bennet, A, Thompson, A.J, Bernardo-Seisdedos, G, Millet, O, Zhu, S, Lu, D, Sollogoub, M, Rovira, C, Jimenez-Barbero, J, Davies, G.J, Williams, S.J. | | Deposit date: | 2018-03-06 | | Release date: | 2019-09-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | An Epoxide Intermediate in Glycosidase Catalysis.
Acs Cent.Sci., 6, 2020
|
|
8DEX
 
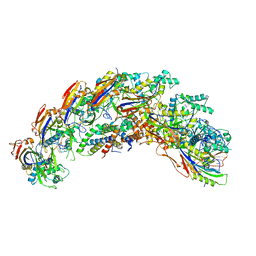 | | type I-C Cascade | | Descriptor: | CRISPR-associated protein, CT1133 family, TM1801 family, ... | | Authors: | O'Brien, R.E, Bravo, J.P.K, Ramos, D, Hibshman, G.N, Wright, J.T, Taylor, D.W. | | Deposit date: | 2022-06-21 | | Release date: | 2023-02-15 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structural snapshots of R-loop formation by a type I-C CRISPR Cascade.
Mol.Cell, 83, 2023
|
|
