7U4W
 
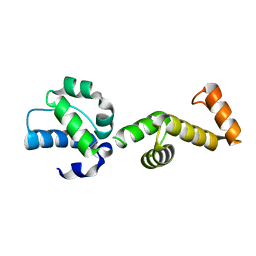 | | The ubiquitin-associated domain of human thirty-eight negative kinase-1 flexibly fused to the 1TEL crystallization chaperone via a 2-glycine linker and crystallized at traditional protein concentration | | Descriptor: | Transcription factor ETV6,Non-receptor tyrosine-protein kinase TNK1 | | Authors: | Nawarathnage, S, Pedroza Romo, M.J, Smith, T, Bunn, D, Stewart, C, Doukov, T, Moody, J.D. | | Deposit date: | 2022-03-01 | | Release date: | 2023-03-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Fusion crystallization reveals the behavior of both the 1TEL crystallization chaperone and the TNK1 UBA domain.
Structure, 31, 2023
|
|
8ANA
 
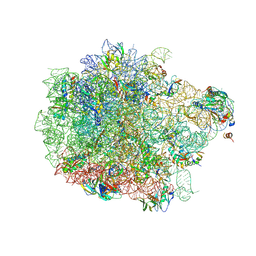 | | Cryo-EM structure of the proline-rich antimicrobial peptide drosocin bound to the 50S ribosomal subunit | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-galactopyranose, 23S ribosomal RNA, 50S ribosomal protein L13, ... | | Authors: | Koller, T.O, Morici, M, Wilson, D.N. | | Deposit date: | 2022-08-05 | | Release date: | 2023-03-08 | | Last modified: | 2023-09-06 | | Method: | ELECTRON MICROSCOPY (2.1 Å) | | Cite: | Structural basis for translation inhibition by the glycosylated drosocin peptide.
Nat.Chem.Biol., 19, 2023
|
|
5L6S
 
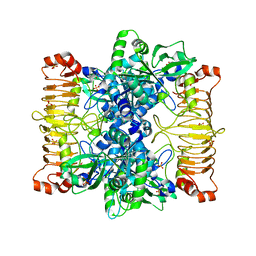 | | Crystal structure of E. coli ADP-glucose pyrophosphorylase (AGPase) in complex with a positive allosteric regulator beta-fructose-1,6-diphosphate (FBP) - AGPase*FBP | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, Glucose-1-phosphate adenylyltransferase, SULFATE ION | | Authors: | Cifuente, J.O, Albesa-Jove, D, Comino, N, Madariaga-Marcos, J, Agirre, J, Lopez-Fernandez, S, Garcia-Alija, M, Guerin, M.E. | | Deposit date: | 2016-05-31 | | Release date: | 2016-09-07 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.04 Å) | | Cite: | Structural Basis of Glycogen Biosynthesis Regulation in Bacteria.
Structure, 24, 2016
|
|
4YW2
 
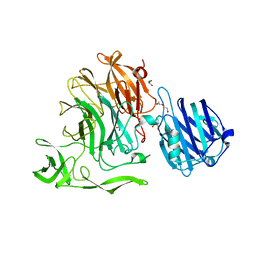 | | Crystal Structure of Streptococcus pneumoniae NanC, complex 6'SL | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, N-acetyl-alpha-neuraminic acid-(2-6)-beta-D-galactopyranose, ... | | Authors: | Owen, C.D, Lukacik, P, Potter, J.A, Walsh, M, Taylor, G.L. | | Deposit date: | 2015-03-20 | | Release date: | 2015-09-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Streptococcus pneumoniae NanC: STRUCTURAL INSIGHTS INTO THE SPECIFICITY AND MECHANISM OF A SIALIDASE THAT PRODUCES A SIALIDASE INHIBITOR.
J.Biol.Chem., 290, 2015
|
|
6G31
 
 | | Crystal structure of human geranylgeranyl diphosphate synthase mutant D188Y bound to zoledronate | | Descriptor: | Geranylgeranyl pyrophosphate synthase, MAGNESIUM ION, ZOLEDRONIC ACID | | Authors: | Lisnyansky, M, Kapelushnik, N, Ben-Bassat, A, Marom, M, Loewenstein, A, Khananshvili, D, Giladi, M, Haitin, Y. | | Deposit date: | 2018-03-24 | | Release date: | 2018-10-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Reduced Activity of Geranylgeranyl Diphosphate Synthase Mutant Is Involved in Bisphosphonate-Induced Atypical Fractures.
Mol. Pharmacol., 94, 2018
|
|
8PVZ
 
 | |
5NLX
 
 | | A2A Adenosine receptor room-temperature structure determined by serial millisecond crystallography | | Descriptor: | 4-{2-[(7-amino-2-furan-2-yl[1,2,4]triazolo[1,5-a][1,3,5]triazin-5-yl)amino]ethyl}phenol, Adenosine receptor A2a,Soluble cytochrome b562,Adenosine receptor A2a, CHOLESTEROL, ... | | Authors: | Weinert, T, Cheng, R, James, D, Gashi, D, Nogly, P, Jaeger, K, Dore, A.S, Geng, T, Cooke, R, Hennig, M, Standfuss, J. | | Deposit date: | 2017-04-05 | | Release date: | 2017-09-27 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Serial millisecond crystallography for routine room-temperature structure determination at synchrotrons.
Nat Commun, 8, 2017
|
|
5NMG
 
 | | 868 TCR in complex with HLA A02 presenting SLYFNTIAVL | | Descriptor: | 1,2-ETHANEDIOL, Beta-2-microglobulin, GLYCEROL, ... | | Authors: | Rizkallah, P.J, Cole, D.K, Fuller, A, Sewell, A.K. | | Deposit date: | 2017-04-05 | | Release date: | 2017-11-15 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Dual Molecular Mechanisms Govern Escape at Immunodominant HLA A2-Restricted HIV Epitope.
Front Immunol, 8, 2017
|
|
6G2U
 
 | | Crystal structure of the human glutamate dehydrogenase 2 (hGDH2) | | Descriptor: | CHLORIDE ION, Glutamate dehydrogenase 2, mitochondrial, ... | | Authors: | Fadouloglou, V.F, Dimovasili, C, Providaki, M, Kotsifaki, D, Sarrou, I, Plaitakis, A, Zaganas, I, Kokkinidis, M. | | Deposit date: | 2018-03-23 | | Release date: | 2019-04-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.934287 Å) | | Cite: | Crystal structure of glutamate dehydrogenase 2, a positively selected novel human enzyme involved in brain biology and cancer pathophysiology.
J.Neurochem., 2021
|
|
6D4M
 
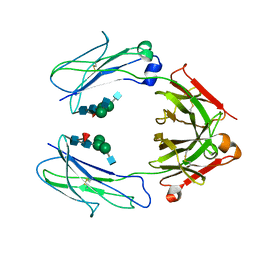 | | Crystal Structure of a Fc Fragment of Rhesus macaque (Macaca mulatta) IgG3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Fc fragment of IgG3 | | Authors: | Gohain, N, Tolbert, W.D, Pazgier, M. | | Deposit date: | 2018-04-18 | | Release date: | 2019-05-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.47 Å) | | Cite: | From Rhesus macaque to human: structural evolutionary pathways for immunoglobulin G subclasses.
Mabs, 11, 2019
|
|
6GJ8
 
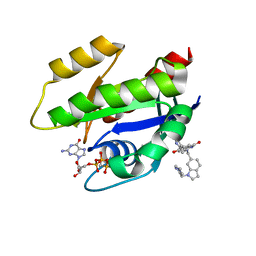 | | CRYSTAL STRUCTURE OF KRAS G12D (GPPCP) IN COMPLEX WITH BI 2852 | | Descriptor: | (3~{S})-3-[2-[[[1-[(1-methylimidazol-4-yl)methyl]indol-6-yl]methylamino]methyl]-1~{H}-indol-3-yl]-5-oxidanyl-2,3-dihydroisoindol-1-one, GTPase KRas, MAGNESIUM ION, ... | | Authors: | Kessler, D, Mcconnell, D.M, Mantoulidis, A. | | Deposit date: | 2018-05-16 | | Release date: | 2019-07-31 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Drugging an undruggable pocket on KRAS.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
7LSA
 
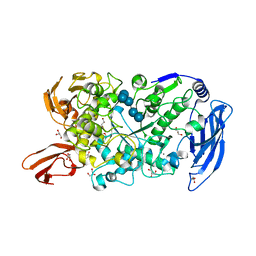 | | Ruminococcus bromii Amy12 with maltoheptaose | | Descriptor: | CALCIUM ION, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Koropatkin, N.M, Cockburn, D.W, Brown, H.A, Kibler, R.D. | | Deposit date: | 2021-02-18 | | Release date: | 2021-07-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Structure and substrate recognition by the Ruminococcus bromii amylosome pullulanases.
J.Struct.Biol., 213, 2021
|
|
7LST
 
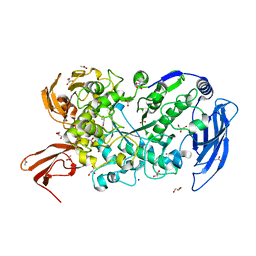 | | Ruminococcus bromii Amy12-D392A with 63-a-D-glucosyl-maltotriosyl-maltotriose | | Descriptor: | ACETATE ION, CALCIUM ION, GLYCEROL, ... | | Authors: | Koropatkin, N.M, Cockburn, D.W, Brown, H.A, Kibler, R.D. | | Deposit date: | 2021-02-18 | | Release date: | 2021-07-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structure and substrate recognition by the Ruminococcus bromii amylosome pullulanases.
J.Struct.Biol., 213, 2021
|
|
6GOS
 
 | | E. coli Microcin synthetase McbBCD complex with pro-MccB17 bound | | Descriptor: | 1,2-ETHANEDIOL, Bacteriocin microcin B17, CHLORIDE ION, ... | | Authors: | Ghilarov, D, Stevenson, C.E.M, Travin, D.Y, Piskunova, J, Serebryakova, M, Maxwell, A, Lawson, D.M, Severinov, K. | | Deposit date: | 2018-06-04 | | Release date: | 2019-01-30 | | Last modified: | 2019-03-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Architecture of Microcin B17 Synthetase: An Octameric Protein Complex Converting a Ribosomally Synthesized Peptide into a DNA Gyrase Poison.
Mol. Cell, 73, 2019
|
|
6T8W
 
 | | Complement factor B in complex with (-)-4-(1-((5,7-Dimethyl-1H-indol-4-yl)methyl)piperidin-2-yl)benzoic acid | | Descriptor: | 5,7-dimethyl-4-[[(2~{S})-2-phenylpiperidin-1-yl]methyl]-1~{H}-indole, Complement factor B, SULFATE ION, ... | | Authors: | Mainolfi, N, Ehara, T, Karki, R.G, Anderson, K, Sweeney, A.M, Wiesmann, C, Adams, C, Mainolfi, N, Liao, S.M, Argikar, U.A, Jendza, K, Zhang, C, Powers, J, Klosowski, D.W, Crowley, M, Kawanami, T, Ding, J, April, M, Forster, C, Wu, M.S, Capparelli, M, Ramqaj, R, Solovay, C, Cumin, F, Smith, T.M, Ferrara, L, Lee, W, Long, D, Prentiss, M, Erkenez, A.D, Yang, L, Fang, L, Sellner, H, Sirockin, F, Valeur, E, Erbel, P, Ramage, P, Gerhartz, B, Schubart, A, Flohr, S, Gradoux, N, Feifel, R, Vogg, B, Wiesmann, C, Maibaum, J, Eder, J, Sedrani, R, Harrison, R.A, Mogi, M, Jaffee, B.D, Adams, C.M. | | Deposit date: | 2019-10-25 | | Release date: | 2020-03-04 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Discovery of 4-((2S,4S)-4-Ethoxy-1-((5-methoxy-7-methyl-1H-indol-4-yl)methyl)piperidin-2-yl)benzoic Acid (LNP023), a Factor B Inhibitor Specifically Designed To Be Applicable to Treating a Diverse Array of Complement Mediated Diseases.
J.Med.Chem., 63, 2020
|
|
7LSU
 
 | | Ruminococcus bromii Amy12-D392A with 63-a-D-glucosyl-maltotriose | | Descriptor: | ACETATE ION, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Koropatkin, N.M, Cockburn, D.W, Brown, H.A, Kibler, R.D. | | Deposit date: | 2021-02-18 | | Release date: | 2021-07-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure and substrate recognition by the Ruminococcus bromii amylosome pullulanases.
J.Struct.Biol., 213, 2021
|
|
7LSR
 
 | | Ruminococcus bromii Amy12-D392A with maltoheptaose | | Descriptor: | CALCIUM ION, GLYCEROL, Pullulanase, ... | | Authors: | Koropatkin, N.M, Cockburn, D.W, Brown, H.A, Kibler, R.D. | | Deposit date: | 2021-02-18 | | Release date: | 2021-07-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Structure and substrate recognition by the Ruminococcus bromii amylosome pullulanases.
J.Struct.Biol., 213, 2021
|
|
5L8G
 
 | | Crystal structure of Rhodospirillum rubrum Rru_A0973 mutant H65A | | Descriptor: | CALCIUM ION, Uncharacterized protein | | Authors: | He, D, Hughes, S, Vanden-Hehir, S, Georgiev, A, Altenbach, K, Tarrant, E, Mackay, C.L, Waldron, K.J, Clarke, D.J, Marles-Wright, J. | | Deposit date: | 2016-06-07 | | Release date: | 2016-08-31 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.974 Å) | | Cite: | Structural characterization of encapsulated ferritin provides insight into iron storage in bacterial nanocompartments.
Elife, 5, 2016
|
|
8ARC
 
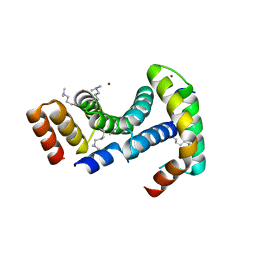 | |
4Z0N
 
 | | Crystal Structure of a Periplasmic Solute binding protein (IPR025997) from Streptobacillus moniliformis DSM-12112 (Smon_0317, TARGET EFI-511281) with bound D-Galactose | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CALCIUM ION, ... | | Authors: | Yadava, U, Vetting, M.W, Al Obaidi, N.F, Toro, R, Morisco, L.L, Benach, J, Koss, J, Wasserman, S.R, Attonito, J.D, Scott Glenn, A, Chamala, S, Chowdhury, S, Lafleur, J, Love, J, Seidel, R.D, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2015-03-26 | | Release date: | 2015-04-15 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.26 Å) | | Cite: | Crystal Structure of a Periplasmic Solute binding protein (IPR025997) from Streptobacillus moniliformis DSM-12112 (Smon_0317, TARGET EFI-511281) with bound D-Galactose
To be published
|
|
6D58
 
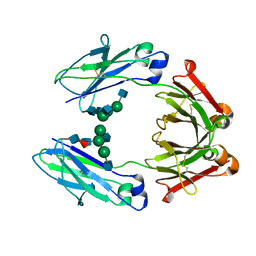 | | Crystal structure of a Fc fragment of Human IgG3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Immunoglobulin heavy constant gamma 3 | | Authors: | Gohain, N, Tolbert, W.D, Pazgier, M. | | Deposit date: | 2018-04-19 | | Release date: | 2019-05-15 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Crystal structure of a Fc fragment of Human IgG3.
To Be Published
|
|
6TDH
 
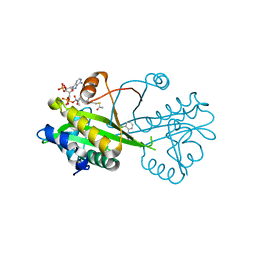 | | Crystal structure of Aspergillus fumigatus Glucosamine-6-phosphate N-acetyltransferase 1 in complex with compound 1 | | Descriptor: | 3-(2-chlorophenyl)-4~{H}-1,2,4-triazole, ACETYL COENZYME *A, Glucosamine 6-phosphate N-acetyltransferase | | Authors: | Raimi, O.G, Stanley, M, Lockhart, D. | | Deposit date: | 2019-11-08 | | Release date: | 2020-04-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Targeting a critical step in fungal hexosamine biosynthesis.
J.Biol.Chem., 295, 2020
|
|
5L8C
 
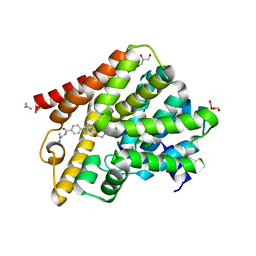 | | Crystal structure of T. brucei PDE-B1 catalytic domain with inhibitor NPD-039 | | Descriptor: | 4-[5-[(4~{a}~{R},8~{a}~{S})-4-oxidanylidene-3-propan-2-yl-4~{a},5,8,8~{a}-tetrahydrophthalazin-1-yl]-2-methoxy-phenyl]-~{N}-(2-azanyl-2-oxidanylidene-ethyl)benzamide, DI(HYDROXYETHYL)ETHER, FORMIC ACID, ... | | Authors: | Singh, A.K, Anthonyrajah, E.S, Brown, D.G. | | Deposit date: | 2016-06-07 | | Release date: | 2018-03-14 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Targeting a Subpocket in Trypanosoma brucei Phosphodiesterase B1 (TbrPDEB1) Enables the Structure-Based Discovery of Selective Inhibitors with Trypanocidal Activity.
J. Med. Chem., 61, 2018
|
|
7KY7
 
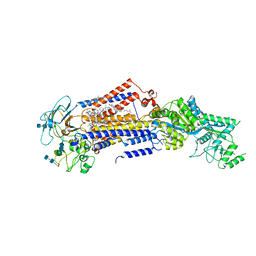 | | Structure of the S. cerevisiae phosphatidylcholine flippase Dnf2-Lem3 complex in the apo E1 state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Alkylphosphocholine resistance protein LEM3, CHOLESTEROL, ... | | Authors: | Bai, L, You, Q, Jain, B.K, Duan, H.D, Kovach, A, Graham, T.R, Li, H. | | Deposit date: | 2020-12-07 | | Release date: | 2021-01-06 | | Method: | ELECTRON MICROSCOPY (3.08 Å) | | Cite: | Transport mechanism of P4 ATPase phosphatidylcholine flippases.
Elife, 9, 2020
|
|
6TKG
 
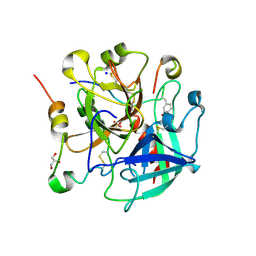 | | Tsetse thrombin inhibitor in complex with human alpha-thrombin - orthorhombic form at 12keV | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, Prothrombin, ... | | Authors: | Calisto, B.M, Ripoll-Rozada, J, de Sanctis, D, Pereira, P.J.B. | | Deposit date: | 2019-11-28 | | Release date: | 2020-11-04 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Sulfotyrosine-Mediated Recognition of Human Thrombin by a Tsetse Fly Anticoagulant Mimics Physiological Substrates.
Cell Chem Biol, 28, 2021
|
|
