6MU3
 
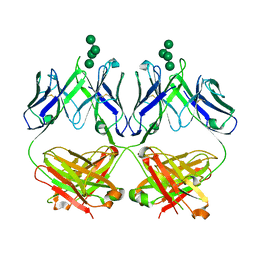 | | Anti-HIV-1 Fab 2G12 + Man7 re-refinement | | Descriptor: | Fab 2G12, heavy chain, light chain, ... | | Authors: | Wilson, I.A, Calarese, D.A, Stanfield, R.L. | | Deposit date: | 2018-10-22 | | Release date: | 2018-10-31 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.327 Å) | | Cite: | Dissection of the carbohydrate specificity of the broadly neutralizing anti-HIV-1 antibody 2G12.
Proc. Natl. Acad. Sci. U.S.A., 102, 2005
|
|
8QEB
 
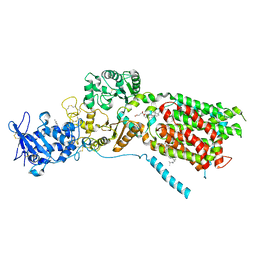 | | S. cerevisia Niemann-Pick type C protein NCR1 in GDN at pH 7.5 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL HEMISUCCINATE, ERGOSTEROL, ... | | Authors: | Frain, K.M, Dedic, E, Nel, L, Olesen, E, Stokes, D, Panyella Pedersen, B. | | Deposit date: | 2023-08-31 | | Release date: | 2023-10-18 | | Last modified: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Conformational changes in the Niemann-Pick type C1 protein NCR1 drive sterol translocation.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8QED
 
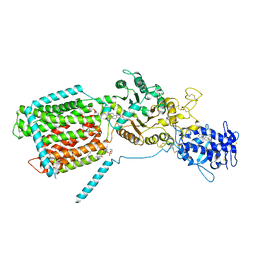 | | S. cerevisia Niemann-Pick type C protein NCR1 in LMNG at pH 5.5 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ERGOSTEROL, NPC intracellular sterol transporter 1-related protein 1, ... | | Authors: | Frain, K.M, Nel, L, Dedic, E, Olesen, E, Stokes, D, Panyella Pedersen, B. | | Deposit date: | 2023-08-31 | | Release date: | 2023-10-18 | | Last modified: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (3.27 Å) | | Cite: | Conformational changes in the Niemann-Pick type C1 protein NCR1 drive sterol translocation.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8QEE
 
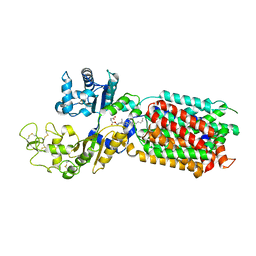 | | S. cerevisia Niemann-Pick type C protein NCR1 in Peptidisc at pH 7.5 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL HEMISUCCINATE, ... | | Authors: | Frain, K.M, Dedic, E, Nel, L, Olesen, E, Stokes, D, Panyella Pedersen, B. | | Deposit date: | 2023-08-31 | | Release date: | 2023-10-18 | | Last modified: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (2.43 Å) | | Cite: | Conformational changes in the Niemann-Pick type C1 protein NCR1 drive sterol translocation.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8FRF
 
 | | Homodimeric designed loop protein RBL7_C2_3 | | Descriptor: | MALONATE ION, RBL7_C2_3 | | Authors: | Jude, K.M, Jiang, H, Baker, D, Garcia, K.C. | | Deposit date: | 2023-01-06 | | Release date: | 2024-04-17 | | Last modified: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | De novo design of buttressed loops for sculpting protein functions.
Nat.Chem.Biol., 20, 2024
|
|
8FYE
 
 | | 4-F, 5-MeO-PyrT-bound serotonin 1A (5-HT1A) receptor-Gi1 protein complex | | Descriptor: | 4-fluoro-5-methoxy-3-[2-(pyrrolidin-1-yl)ethyl]-1H-indole, CHOLESTEROL HEMISUCCINATE, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Warren, A.L, Zilberg, G, Capper, M.J, Wacker, D. | | Deposit date: | 2023-01-26 | | Release date: | 2024-05-15 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.85 Å) | | Cite: | Structural pharmacology and therapeutic potential of 5-methoxytryptamines.
Nature, 630, 2024
|
|
5NDH
 
 | | The structure of the G. violaceus guanidine II riboswitch P2 stem-loop | | Descriptor: | GUANIDINE, MAGNESIUM ION, RNA (5'-R(*GP*(CBV)P*GP*GP*GP*GP*AP*CP*GP*AP*CP*CP*CP*CP*GP*C)-3'), ... | | Authors: | Huang, L, Wang, J, Lilley, D.M.J. | | Deposit date: | 2017-03-08 | | Release date: | 2017-05-31 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | The Structure of the Guanidine-II Riboswitch.
Cell Chem Biol, 24, 2017
|
|
8FYT
 
 | | LSD-bound serotonin 1A (5-HT1A) receptor-Gi1 protein complex | | Descriptor: | (8alpha)-N,N-diethyl-6-methyl-9,10-didehydroergoline-8-carboxamide, CHOLESTEROL HEMISUCCINATE, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Warren, A.L, Zilberg, G, Capper, M.J, Wacker, D. | | Deposit date: | 2023-01-26 | | Release date: | 2024-05-15 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.64 Å) | | Cite: | Structural pharmacology and therapeutic potential of 5-methoxytryptamines.
Nature, 630, 2024
|
|
6WB1
 
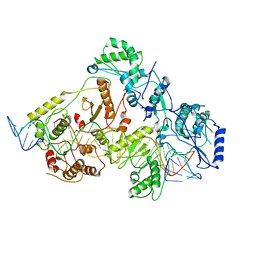 | | +3 extended HIV-1 reverse transcriptase initiation complex core (intermediate state) | | Descriptor: | HIV-1 viral RNA genome fragment, REVERSE TRANSCRIPTASE/RIBONUCLEASE H, reverse transcriptase p51 subunit, ... | | Authors: | Larsen, K.P, Jackson, L.N, Kappel, K, Zhang, J, Chen, D.H, Puglisi, E.V. | | Deposit date: | 2020-03-26 | | Release date: | 2020-06-24 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | Distinct Conformational States Underlie Pausing during Initiation of HIV-1 Reverse Transcription.
J.Mol.Biol., 432, 2020
|
|
8FYL
 
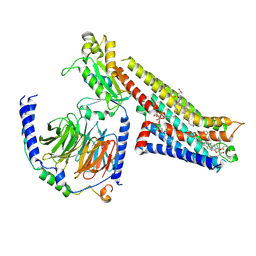 | | Vilazodone-bound serotonin 1A (5-HT1A) receptor-Gi1 protein complex | | Descriptor: | 5-{4-[4-(5-cyano-1H-indol-3-yl)butyl]piperazin-1-yl}-1-benzofuran-2-carboxamide, CHOLESTEROL HEMISUCCINATE, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Warren, A.L, Zilberg, G, Capper, M.J, Wacker, D. | | Deposit date: | 2023-01-26 | | Release date: | 2024-05-15 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.94 Å) | | Cite: | Structural pharmacology and therapeutic potential of 5-methoxytryptamines.
Nature, 630, 2024
|
|
7QXF
 
 | |
7QOF
 
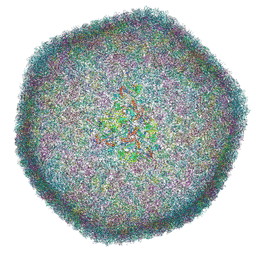 | | Icosahedral capsid of the phicrAss001 virion | | Descriptor: | Auxiliary capsid protein gp36, Head fiber dimer protein gp29, Head fiber trimer protein gp21, ... | | Authors: | Bayfield, O.W, Shkoporov, A.N, Yutin, N, Khokhlova, E.V, Smith, J.L.R, Hawkins, D.E.D.P, Koonin, E.V, Hill, C, Antson, A.A. | | Deposit date: | 2021-12-24 | | Release date: | 2023-03-29 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.01 Å) | | Cite: | Structural atlas of a human gut crassvirus.
Nature, 617, 2023
|
|
7QOH
 
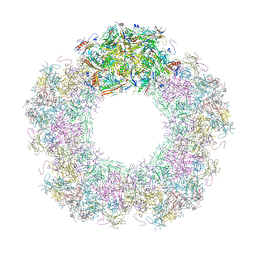 | | Unique vertex of the phicrAss001 virion with C5 symmetry imposed | | Descriptor: | Auxiliary capsid protein gp36, Head fiber trimer protein gp21, MAGNESIUM ION, ... | | Authors: | Bayfield, O.W, Shkoporov, A.N, Yutin, N, Khokhlova, E.V, Smith, J.L.R, Hawkins, D.E.D.P, Koonin, E.V, Hill, C, Antson, A.A. | | Deposit date: | 2021-12-24 | | Release date: | 2023-03-29 | | Last modified: | 2023-05-24 | | Method: | ELECTRON MICROSCOPY (3.32 Å) | | Cite: | Structural atlas of a human gut crassvirus.
Nature, 617, 2023
|
|
7QOK
 
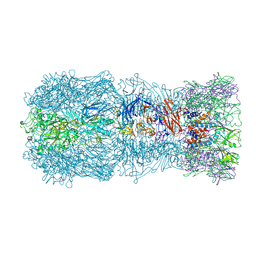 | | Tail muzzle assembly of the phicrAss001 virion with C6 symmetry imposed | | Descriptor: | MAGNESIUM ION, Muzzle bound helix, Muzzle protein gp44, ... | | Authors: | Bayfield, O.W, Shkoporov, A.N, Yutin, N, Khokhlova, E.V, Smith, J.L.R, Hawkins, D.E.D.P, Koonin, E.V, Hill, C, Antson, A.A. | | Deposit date: | 2021-12-24 | | Release date: | 2023-03-29 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.38 Å) | | Cite: | Structural atlas of a human gut crassvirus.
Nature, 617, 2023
|
|
7QOL
 
 | | Tail assembly of the phicrAss001 virion with C6 symmetry imposed | | Descriptor: | Cargo protein 1 gp45, MAGNESIUM ION, Muzzle bound helix, ... | | Authors: | Bayfield, O.W, Shkoporov, A.N, Yutin, N, Khokhlova, E.V, Smith, J.L.R, Hawkins, D.E.D.P, Koonin, E.V, Hill, C, Antson, A.A. | | Deposit date: | 2021-12-24 | | Release date: | 2023-03-29 | | Last modified: | 2023-05-24 | | Method: | ELECTRON MICROSCOPY (3.33 Å) | | Cite: | Structural atlas of a human gut crassvirus.
Nature, 617, 2023
|
|
7OR6
 
 | |
6W1U
 
 | | RT XFEL structure of Photosystem II 400 microseconds after the second illumination at 2.09 Angstrom resolution | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Ibrahim, M, Fransson, T, Chatterjee, R, Cheah, M.H, Hussein, R, Lassalle, L, Sutherlin, K.D, Young, I.D, Fuller, F.D, Gul, S, Kim, I.-S, Simon, P.S, de Lichtenberg, C, Chernev, P, Bogacz, I, Pham, C, Orville, A.M, Saichek, N, Northen, T.R, Batyuk, A, Carbajo, S, Alonso-Mori, R, Tono, K, Owada, S, Bhowmick, A, Bolotovski, R, Mendez, D, Moriarty, N.W, Holton, J.M, Dobbek, H, Brewster, A.S, Adams, P.D, Sauter, N.K, Bergmann, U, Zouni, A, Messinger, J, Kern, J, Yachandra, V.K, Yano, J. | | Deposit date: | 2020-03-04 | | Release date: | 2020-05-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Untangling the sequence of events during the S2→ S3transition in photosystem II and implications for the water oxidation mechanism.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
7ORD
 
 | |
8TTZ
 
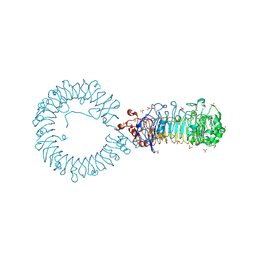 | | Crystal structure of monkey TLR7 ectodomain with compound 20 | | Descriptor: | (3S)-3-({5-amino-1-[(2-methoxy-4-{[(oxan-4-yl)amino]methyl}phenyl)methyl]-1H-pyrazolo[4,3-d]pyrimidin-7-yl}amino)hexan-1-ol, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Critton, D.A. | | Deposit date: | 2023-08-15 | | Release date: | 2024-02-07 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.14 Å) | | Cite: | Discovery of Novel TLR7 Agonists as Systemic Agent for Combination With aPD1 for Use in Immuno-oncology.
Acs Med.Chem.Lett., 15, 2024
|
|
6Z84
 
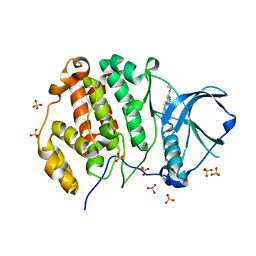 | | CK2 alpha bound to chemical probe SGC-CK2-1 derivative | | Descriptor: | Casein kinase II subunit alpha, SULFATE ION, ~{N}-[1-[3-cyano-7-(cyclopropylamino)pyrazolo[1,5-a]pyrimidin-5-yl]indol-6-yl]ethanamide | | Authors: | Kraemer, A, Wells, C, Drewry, D.H, Pickett, J.E, Axtman, A.D, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-06-02 | | Release date: | 2020-07-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Development of a potent and selective chemical probe for the pleiotropic kinase CK2.
Cell Chem Biol, 28, 2021
|
|
7OWC
 
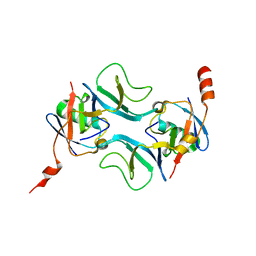 | |
7TUF
 
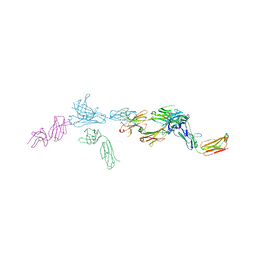 | | Crystal structure of Tapasin in complex with PaSta1-Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, PaSta1 Fab heavy chain, PaSta1 Fab kappa light chain, ... | | Authors: | Jiang, J, Natarajan, K, Taylor, D.K, Boyd, L.F, Margulies, D.H. | | Deposit date: | 2022-02-02 | | Release date: | 2022-09-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural mechanism of tapasin-mediated MHC-I peptide loading in antigen presentation.
Nat Commun, 13, 2022
|
|
8UPV
 
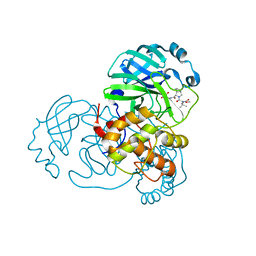 | | Structure of SARS-Cov2 3CLPro in complex with Compound 33 | | Descriptor: | 3C-like proteinase nsp5, methyl {(2S)-1-[(1S,3aR,6aS)-1-{[(2R,3S,6R)-6-fluoro-2-hydroxy-1-(methylamino)-1-oxoheptan-3-yl]carbamoyl}hexahydrocyclopenta[c]pyrrol-2(1H)-yl]-3,3-dimethyl-1-oxobutan-2-yl}carbamate | | Authors: | Krishnamurthy, H, Zhuang, N, Qiang, D, Wu, Y, Klein, D.J. | | Deposit date: | 2023-10-23 | | Release date: | 2024-03-06 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Invention of MK-7845, a SARS-CoV-2 3CL Protease Inhibitor Employing a Novel Difluorinated Glutamine Mimic.
J.Med.Chem., 67, 2024
|
|
8G45
 
 | | Structure of HDAC6 zinc-finger ubiquitin binding domain in complex with SGC-UBD253 chemical probe | | Descriptor: | 3-[8-chloro-3-(2-{[(2-methoxyphenyl)methyl]amino}-2-oxoethyl)-4-oxo-3,4-dihydroquinazolin-2-yl]propanoic acid, Histone deacetylase 6, ZINC ION | | Authors: | Harding, R.J, Franzoni, I, Mann, M.K, Szewczyk, M, Mirabi, B, Owens, D.D.G, Ackloo, S, Scheremetjew, A, Juarez-Ornelas, K.A, Sanichar, R, Baker, R.J, Dank, C, Brown, P.J, Barsyte-Lovejoy, D, Santhakumar, V, Schapira, M, Lautens, M, Arrowsmith, C.H, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-02-08 | | Release date: | 2023-05-03 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Discovery and Characterization of a Chemical Probe Targeting the Zinc-Finger Ubiquitin-Binding Domain of HDAC6.
J.Med.Chem., 66, 2023
|
|
8G43
 
 | | Structure of HDAC6 zinc-finger ubiquitin binding domain in complex with 3-(3-(2-(methylamino)-2-oxoethyl)-4-oxo-3,4-dihydroquinazolin-2-yl)propanoic acid | | Descriptor: | 3-{3-[2-(methylamino)-2-oxoethyl]-4-oxo-3,4-dihydroquinazolin-2-yl}propanoic acid, Histone deacetylase 6, ZINC ION | | Authors: | Harding, R.J, Franzoni, I, Mann, M.K, Szewczyk, M, Mirabi, B, Owens, D.D.G, Ackloo, S, Scheremetjew, A, Juarez-Ornelas, K.A, Sanichar, R, Baker, R.J, Dank, C, Brown, P.J, Barsyte-Lovejoy, D, Santhakumar, V, Schapira, M, Lautens, M, Arrowsmith, C.H, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-02-08 | | Release date: | 2023-05-03 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Discovery and Characterization of a Chemical Probe Targeting the Zinc-Finger Ubiquitin-Binding Domain of HDAC6.
J.Med.Chem., 66, 2023
|
|
