4IO8
 
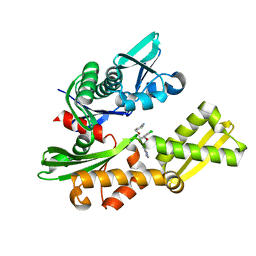 | | Crystal structure of human HSP70 complexed with 4-{(2R,3S,4R)-5-[(R)-6-Amino-8-(3,4-dichloro-benzylamino)-purin-9-yl]-3,4-dihydroxy-tetrahydro-furan-2-ylmethoxymethyl}-benzonitrile | | Descriptor: | 4-[[(2R,3S,4R,5R)-5-[6-amino-8-[(3,4-dichlorophenyl)methylamino]purin-9-yl]-3,4-dihydroxy-oxolan-2-yl]methoxymethyl]benzonitrile, Heat shock 70kDa protein 1A variant | | Authors: | Musil, D, Scholz, S. | | Deposit date: | 2013-01-07 | | Release date: | 2013-12-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Functional analysis of hsp70 inhibitors.
Plos One, 8, 2013
|
|
7T3X
 
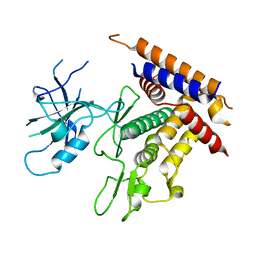 | | Structure of unphosphorylated Pediculus humanus (Ph) PINK1 D334A mutant | | Descriptor: | Serine/threonine-protein kinase PINK1 | | Authors: | Gan, Z.Y, Leis, A, Dewson, G, Glukhova, A, Komander, D. | | Deposit date: | 2021-12-09 | | Release date: | 2021-12-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.53 Å) | | Cite: | Activation mechanism of PINK1.
Nature, 602, 2022
|
|
7T44
 
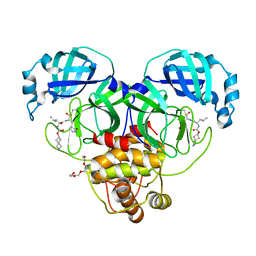 | | Structure of SARS-CoV-2 3CL protease in complex with inhibitor 4c | | Descriptor: | (1R,2S)-2-[(N-{[(2-azaspiro[3.3]heptan-6-yl)oxy]carbonyl}-L-leucyl)amino]-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, (1S,2S)-1-hydroxy-2-{[N-({[2-(methanesulfonyl)-2-azaspiro[3.3]heptan-6-yl]oxy}carbonyl)-L-leucyl]amino}-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-like proteinase, ... | | Authors: | Liu, L, Lovell, S, Battaile, K.P, Chamandi, S.D, Kim, Y, Groutas, W.C, Chang, K.O. | | Deposit date: | 2021-12-09 | | Release date: | 2021-12-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structure-Guided Design of Potent Spirocyclic Inhibitors of Severe Acute Respiratory Syndrome Coronavirus-2 3C-like Protease.
J.Med.Chem., 65, 2022
|
|
3EFO
 
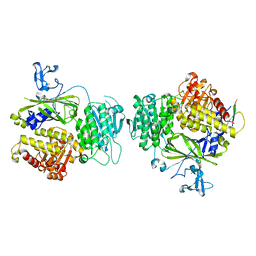 | |
4R60
 
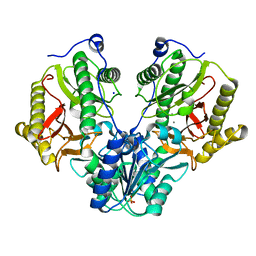 | | Crystal Structure of Xaa-Pro dipeptidase from Xanthomonas campestris | | Descriptor: | MANGANESE (II) ION, PHOSPHATE ION, Proline dipeptidase, ... | | Authors: | Kumar, A, Ghosh, B, Are, V.N, Jamdar, S.N, Makde, R.D, Sharma, S.M. | | Deposit date: | 2014-08-22 | | Release date: | 2014-09-17 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Crystal Structure of Xaa-Pro dipeptidase from Xanthomonas campestris
to be published
|
|
2IAH
 
 | | Crystal structure of the ferripyoverdine receptor of the outer membrane of Pseudomonas aeruginosa bound to ferripyoverdine. | | Descriptor: | (1S)-1-CARBOXY-5-[(3-CARBOXYPROPANOYL)AMINO]-8,9-DIHYDROXY-1,2,3,4-TETRAHYDROPYRIMIDO[1,2-A]QUINOLIN-11-IUM, FE (III) ION, Ferripyoverdine receptor, ... | | Authors: | Wirth, C, Pattus, F, Cobessi, D. | | Deposit date: | 2006-09-08 | | Release date: | 2007-09-11 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | From the periplasmic signaling domain to the extracellular face of an outer membrane signal transducer of Pseudomonas aeruginosa: crystal structure of the ferric pyoverdine outer membrane receptor.
J.Mol.Biol., 368, 2007
|
|
6W2Q
 
 | | Junction 34, DHR53-DHR4 | | Descriptor: | CALCIUM ION, Junction 34 | | Authors: | Bick, M.J, Brunette, T.J, Baker, D. | | Deposit date: | 2020-03-08 | | Release date: | 2020-04-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Modular repeat protein sculpting using rigid helical junctions.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
7T4K
 
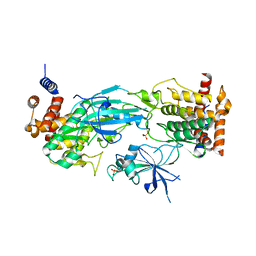 | | Structure of dimeric phosphorylated Pediculus humanus (Ph) PINK1 with kinked alpha-C helix in chain B | | Descriptor: | Serine/threonine-protein kinase PINK1, putative | | Authors: | Gan, Z.Y, Leis, A, Dewson, G, Glukhova, A, Komander, D. | | Deposit date: | 2021-12-10 | | Release date: | 2022-01-12 | | Last modified: | 2022-02-23 | | Method: | ELECTRON MICROSCOPY (3.25 Å) | | Cite: | Activation mechanism of PINK1.
Nature, 602, 2022
|
|
6W3D
 
 | | Rd1NTF2_05 with long sheet | | Descriptor: | Rd1NTF2_05 | | Authors: | Bick, M.J, Basanta, B, Sankaran, B, Baker, D. | | Deposit date: | 2020-03-09 | | Release date: | 2020-04-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | An enumerative algorithm for de novo design of proteins with diverse pocket structures.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
7T4N
 
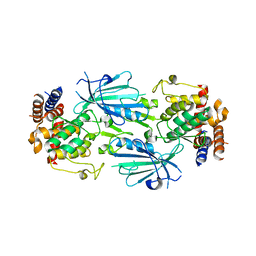 | | Structure of dimeric unphosphorylated Pediculus humanus (Ph) PINK1 D357A mutant | | Descriptor: | Serine/threonine-protein kinase PINK1, putative | | Authors: | Gan, Z.Y, Leis, A, Dewson, G, Glukhova, A, Komander, D. | | Deposit date: | 2021-12-10 | | Release date: | 2022-01-12 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (2.35 Å) | | Cite: | Activation mechanism of PINK1.
Nature, 602, 2022
|
|
6W2W
 
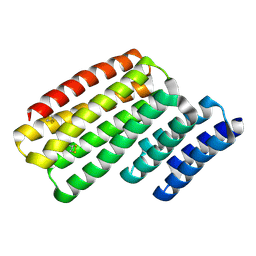 | | Junction 24, DHR14-DHR18 | | Descriptor: | Junction 24 DHR14-DHR18 | | Authors: | Bick, M.J, Brunette, T.J, Baker, D. | | Deposit date: | 2020-03-08 | | Release date: | 2020-04-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Modular repeat protein sculpting using rigid helical junctions.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
4QXP
 
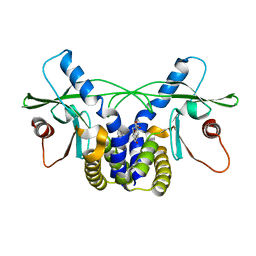 | | Crystal structure of hSTING(G230I) in complex with DMXAA | | Descriptor: | (5,6-dimethyl-9-oxo-9H-xanthen-4-yl)acetic acid, Stimulator of interferon genes protein | | Authors: | Gao, P, Patel, D.J. | | Deposit date: | 2014-07-21 | | Release date: | 2014-09-10 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Binding-Pocket and Lid-Region Substitutions Render Human STING Sensitive to the Species-Specific Drug DMXAA.
Cell Rep, 8, 2014
|
|
7T4L
 
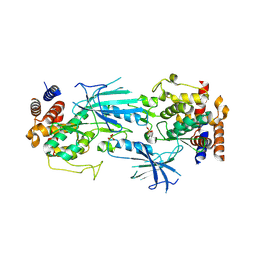 | | Structure of dimeric phosphorylated Pediculus humanus (Ph) PINK1 with extended alpha-C helix in chain B | | Descriptor: | Serine/threonine-protein kinase PINK1, putative | | Authors: | Gan, Z.Y, Leis, A, Dewson, G, Glukhova, A, Komander, D. | | Deposit date: | 2021-12-10 | | Release date: | 2022-01-12 | | Last modified: | 2022-02-23 | | Method: | ELECTRON MICROSCOPY (3.28 Å) | | Cite: | Activation mechanism of PINK1.
Nature, 602, 2022
|
|
6W9Q
 
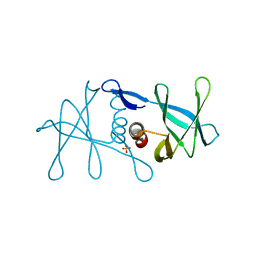 | | Peptide-bound SARS-CoV-2 Nsp9 RNA-replicase | | Descriptor: | 3C-like proteinase peptide, Non-structural protein 9 fusion, PHOSPHATE ION | | Authors: | Littler, D.R, Gully, B.S, Riboldi-Tunnicliffe, A, Rossjohn, J. | | Deposit date: | 2020-03-23 | | Release date: | 2020-04-08 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal Structure of the SARS-CoV-2 Non-structural Protein 9, Nsp9.
Iscience, 23, 2020
|
|
6ITS
 
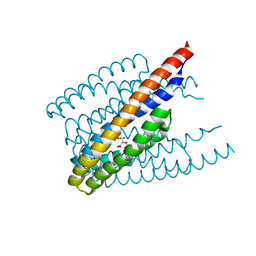 | |
7TB6
 
 | | Structure of S. maltophilia CapW | | Descriptor: | S. maltophilia CapW, SULFATE ION | | Authors: | Blankenchip, C.L, Nguyen, J.V, Lau, R.K, Ye, Q, Corbett, K.D. | | Deposit date: | 2021-12-21 | | Release date: | 2022-01-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Control of bacterial immune signaling by a WYL domain transcription factor.
Nucleic Acids Res., 50, 2022
|
|
7TB5
 
 | | Structure of P. aeruginosa PA17 CapW | | Descriptor: | SULFATE ION, WYL domain-containing protein | | Authors: | Blankenchip, C.L, Nguyen, J.V, Lau, R.K, Ye, Q, Corbett, K.D. | | Deposit date: | 2021-12-21 | | Release date: | 2022-01-19 | | Last modified: | 2022-06-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Control of bacterial immune signaling by a WYL domain transcription factor.
Nucleic Acids Res., 50, 2022
|
|
3ZRI
 
 | |
4QQS
 
 | | Crystal structure of a thermostable family-43 glycoside hydrolase | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Glycoside hydrolase family 43, SODIUM ION | | Authors: | Hassan, N, Kori, L.D, Patel, B.K.C, Divne, C, Tan, T.C. | | Deposit date: | 2014-06-29 | | Release date: | 2015-03-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | High-resolution crystal structure of a polyextreme GH43 glycosidase from Halothermothrix orenii with alpha-L-arabinofuranosidase activity.
Acta Crystallogr F Struct Biol Commun, 71, 2015
|
|
4QPP
 
 | | The Crystal Structure of Human HMT1 hnRNP methyltransferase-like protein 6 in complex with compound DS-421 (2-{4-[3-CHLORO-2-(2-METHOXYPHENYL)-1H-INDOL-5-YL]PIPERIDIN-1-YL}-N-METHYLETHANAMINE | | Descriptor: | 2-{4-[3-chloro-2-(2-methoxyphenyl)-1H-indol-5-yl]piperidin-1-yl}-N-methylethanamine, POLY-UNK, Protein arginine N-methyltransferase 6, ... | | Authors: | Dong, A, Zeng, H, Smil, D, Walker, J.R, He, H, Eram, M, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Vedadi, M, Brown, P.J, Wu, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2014-06-24 | | Release date: | 2014-08-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | The Crystal Structure of Human HMT1
hnRNP methyltransferase-like protein 6 in complex with compound DS-421
To be Published
|
|
2HIP
 
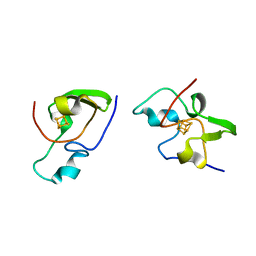 | | THE MOLECULAR STRUCTURE OF THE HIGH POTENTIAL IRON-SULFUR PROTEIN ISOLATED FROM ECTOTHIORHODOSPIRA HALOPHILA DETERMINED AT 2.5-ANGSTROMS RESOLUTION | | Descriptor: | HIGH POTENTIAL IRON SULFUR PROTEIN, IRON/SULFUR CLUSTER | | Authors: | Breiter, D.R, Meyer, T.E, Rayment, I, Holden, H.M. | | Deposit date: | 1991-06-24 | | Release date: | 1992-07-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The molecular structure of the high potential iron-sulfur protein isolated from Ectothiorhodospira halophila determined at 2.5-A resolution.
J.Biol.Chem., 266, 1991
|
|
6IFF
 
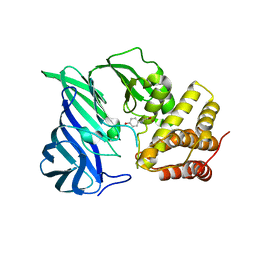 | | Crystal structure of M1 zinc metallopeptidase E323A mutant from Deinococcus radiodurans | | Descriptor: | SODIUM ION, TYROSINE, ZINC ION, ... | | Authors: | Agrawal, R, Kumar, A, Kumar, A, Gaur, N.K, Makde, R.D. | | Deposit date: | 2018-09-20 | | Release date: | 2019-09-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Structural basis for the unusual substrate specificity of unique two-domain M1 metallopeptidase.
Int.J.Biol.Macromol., 147, 2020
|
|
7TGG
 
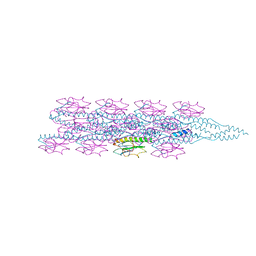 | | Cryo-EM structure of PilA-N and PilA-C from Geobacter sulfurreducens | | Descriptor: | Geopilin domain 1 protein, Geopilin domain 2 protein | | Authors: | Wang, F, Mustafa, K, Chan, C.H, Joshi, K, Bond, D.R, Hochbaum, A.I, Egelman, E.H. | | Deposit date: | 2022-01-07 | | Release date: | 2022-02-23 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Cryo-EM structure of an extracellular Geobacter OmcE cytochrome filament reveals tetrahaem packing.
Nat Microbiol, 7, 2022
|
|
6PQA
 
 | |
4R04
 
 | |
