2XLY
 
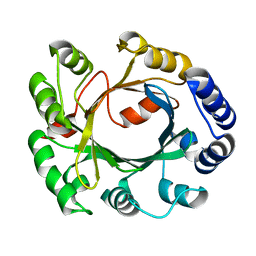 | | Structural and Mechanistic Analysis of the Magnesium-Independent Aromatic Prenyltransferase CloQ from the Clorobiocin Biosynthetic Pathway | | Descriptor: | CLOQ | | Authors: | Metzger, U, Keller, S, Stevenson, C.E.M, Heide, L, Lawson, D.M. | | Deposit date: | 2010-07-22 | | Release date: | 2010-10-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure and Mechanism of the Magnesium-Independent Aromatic Prenyltransferase Cloq from the Clorobiocin Biosynthetic Pathway.
J.Mol.Biol., 404, 2010
|
|
1MJN
 
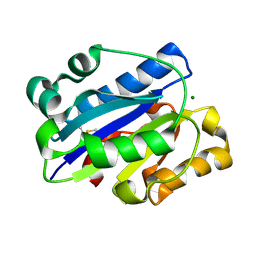 | | Crystal Structure of the intermediate affinity aL I domain mutant | | Descriptor: | Integrin alpha-L, MAGNESIUM ION | | Authors: | Shimaoka, M, Xiao, T, Liu, J.H, Yang, Y.T, Dong, Y.C, Jun, C.D, McCormack, A, Zhang, R.G, Wang, J.H, Springer, T.A. | | Deposit date: | 2002-08-28 | | Release date: | 2003-01-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structures of the alphaL I Domain and its Complex with ICAM-1 reveal a Shape-shifting Pathway for Integrin Regulation
Cell(Cambridge,Mass.), 112, 2003
|
|
2Y0A
 
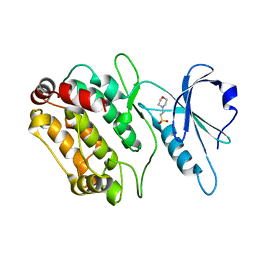 | | Structure of DAPK1 construct residues 1-304 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, DEATH-ASSOCIATED PROTEIN KINASE 1 | | Authors: | Yumerefendi, H, Mas, P.J, Dordevic, N, McCarthy, A.A, Hart, D.J. | | Deposit date: | 2010-12-01 | | Release date: | 2011-12-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Library-Based Construct Screening of Death-Associated Protein Kinase 1 Identifies the Minimal Calmodulin Interaction Region and Autoinhibitory Conformation of the Catalytic Domain
To be Published
|
|
2A0T
 
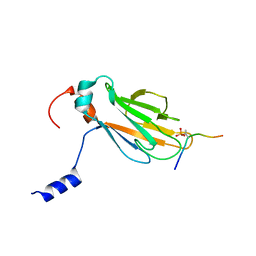 | | NMR structure of the FHA1 domain of Rad53 in complex with a biological relevant phosphopeptide derived from Madt1 | | Descriptor: | Hypothetical 73.8 kDa protein in SAS3-SEC17 intergenic region, residues 301-310, Serine/threonine-protein kinase RAD53 | | Authors: | Mahajan, A, Yuan, C, Pike, B.L, Heierhorst, J, Chang, C.-F, Tsai, M.-D. | | Deposit date: | 2005-06-16 | | Release date: | 2005-11-08 | | Last modified: | 2024-11-13 | | Method: | SOLUTION NMR | | Cite: | FHA Domain-Ligand Interactions: Importance of Integrating Chemical and Biological Approaches
J.Am.Chem.Soc., 127, 2005
|
|
1NJ3
 
 | | Structure and Ubiquitin Interactions of the Conserved NZF Domain of Npl4 | | Descriptor: | NPL4, ZINC ION | | Authors: | Wang, B, Alam, S.L, Meyer, H.H, Payne, M, Stemmler, T.L, Davis, D.R, Sundquist, W.I. | | Deposit date: | 2002-12-30 | | Release date: | 2003-04-22 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure and ubiquitin interactions of the conserved zinc finger domain of Npl4.
J.Biol.Chem., 278, 2003
|
|
2R18
 
 | | Structural insights into the multifunctional protein VP3 of Birnaviruses | | Descriptor: | Capsid assembly protein VP3 | | Authors: | Casanas, A, Navarro, A, Ferrer-Orta, C, Gonzalez, D, Rodriguez, J.F, Verdaguer, N. | | Deposit date: | 2007-08-22 | | Release date: | 2008-02-05 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural insights into the multifunctional protein VP3 of birnaviruses.
Structure, 16, 2008
|
|
1NAF
 
 | |
2G9N
 
 | | Structure of the DEAD domain of Human eukaryotic initiation factor 4A, eIF4A | | Descriptor: | Eukaryotic initiation factor 4A-I | | Authors: | Hogbom, M, Ogg, D, Arrowsmith, C, Berglund, H, Collins, R, Edwards, A, Ehn, M, Flodin, S, Flores, A, Graslund, S, Hallberg, B.M, Hammarstrom, M, Kotenyova, T, Nilsson-Ehle, P, Nordlund, P, Nyman, T, Persson, C, Sagemark, J, Stenmark, P, Sundstrom, M, Thorsell, A.G, Uppenberg, J, Van Den Berg, S, Weigelt, J, Holmberg-Schiavone, L, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-03-07 | | Release date: | 2006-03-14 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Comparative Structural Analysis of Human DEAD-Box RNA Helicases.
Plos One, 5, 2010
|
|
2QVX
 
 | | 4-Chlorobenzoyl-CoA Ligase/Synthetase, I303G mutation, bound to 3-Chlorobenzoate | | Descriptor: | 3-chlorobenzoate, 4-Chlorobenzoate CoA Ligase | | Authors: | Wu, R, Reger, A.S, Cao, J, Gulick, A.M, Dunaway-Mariano, D. | | Deposit date: | 2007-08-09 | | Release date: | 2007-12-18 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Rational redesign of the 4-chlorobenzoate binding site of 4-chlorobenzoate: coenzyme a ligase for expanded substrate range.
Biochemistry, 46, 2007
|
|
5R61
 
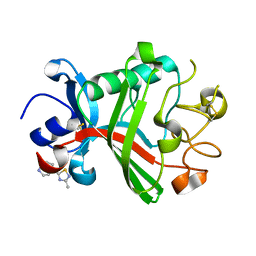 | | PanDDA analysis group deposition -- Crystal Structure of FIBRINOGEN-LIKE GLOBE DOMAIN OF HUMAN TENASCIN-C in complex with Z1578665941 | | Descriptor: | 1-(3-methyl-1,2,4-thiadiazol-5-yl)-1,4-diazepane, Tenascin C (Hexabrachion), isoform CRA_a | | Authors: | Coker, J.A, Bezerra, G.A, von Delft, F, Arrowsmith, C.H, Bountra, C, Edwards, A.M, Yue, W.W, Marsden, B.D. | | Deposit date: | 2020-02-28 | | Release date: | 2020-10-28 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
4B2M
 
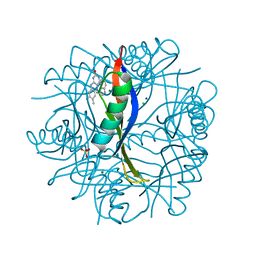 | | COMPLEXES OF DODECIN WITH FLAVIN AND FLAVIN-LIKE LIGANDS | | Descriptor: | CHLORIDE ION, DODECIN, MAGNESIUM ION, ... | | Authors: | Staudt, H, Hoesl, M, Dreuw, A, Serdjukow, S, Oesterhelt, D, Budisa, N, Wachtveitl, J, Grininger, M. | | Deposit date: | 2012-07-16 | | Release date: | 2013-05-29 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The flavoprotein dodecin as a redox probe for electron transfer through DNA.
Angew.Chem.Int.Ed.Engl., 52, 2013
|
|
2AZQ
 
 | | Crystal Structure of Catechol 1,2-Dioxygenase from Pseudomonas arvilla C-1 | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSHOCHOLINE, FE (III) ION, catechol 1,2-dioxygenase | | Authors: | Earhart, C.A, Vetting, M.W, Gosu, R, Michaud-Soret, I, Que, L, Ohlendorf, D.H. | | Deposit date: | 2005-09-12 | | Release date: | 2005-10-04 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structure of catechol 1,2-dioxygenase from Pseudomonas arvilla
Biochem.Biophys.Res.Commun., 338, 2005
|
|
1NH1
 
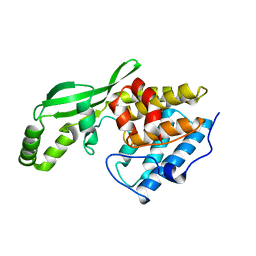 | | Crystal Structure of the Type III Effector AvrB from Pseudomonas syringae. | | Descriptor: | Avirulence B protein | | Authors: | Lee, C.C, Wood, M.D, Ng, K, Luginbuhl, P, Spraggon, G, Katagiri, F. | | Deposit date: | 2002-12-18 | | Release date: | 2004-03-09 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of the Type III Effector AvrB from Pseudomonas syringae.
Structure, 12, 2004
|
|
5R5V
 
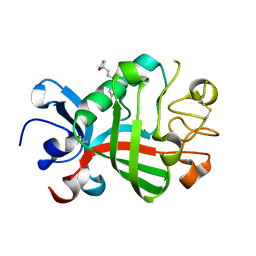 | | PanDDA analysis group deposition -- Crystal Structure of FIBRINOGEN-LIKE GLOBE DOMAIN OF HUMAN TENASCIN-C in complex with Z2856434824 | | Descriptor: | 1-(4-fluorophenyl)-~{N}-[[(2~{R})-oxolan-2-yl]methyl]methanamine, Tenascin C (Hexabrachion), isoform CRA_a | | Authors: | Coker, J.A, Bezerra, G.A, von Delft, F, Arrowsmith, C.H, Bountra, C, Edwards, A.M, Yue, W.W, Marsden, B.D. | | Deposit date: | 2020-02-28 | | Release date: | 2020-10-28 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
1N7P
 
 | | Streptococcus pneumoniae Hyaluronate Lyase W292A/F343V Double Mutant | | Descriptor: | HYALURONIDASE | | Authors: | Nukui, M, Taylor, K.B, McPherson, D.T, Shigenaga, M, Jedrzejas, M.J. | | Deposit date: | 2002-11-16 | | Release date: | 2002-12-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | The function of hydrophobic residues in the catalytic cleft of Streptococcus
pneumoniae hyaluronate lyase. Kinetic characterization of mutant enzyme forms
J.Biol.Chem., 278, 2003
|
|
2PAC
 
 | | SOLUTION STRUCTURE OF FE(II) CYTOCHROME C551 FROM PSEUDOMONAS AERUGINOSA AS DETERMINED BY TWO-DIMENSIONAL 1H NMR | | Descriptor: | CYTOCHROME C551, HEME C | | Authors: | Detlefsen, D.J, Thanabal, V, Pecoraro, V.L, Wagner, G. | | Deposit date: | 1993-05-05 | | Release date: | 1993-10-31 | | Last modified: | 2024-10-09 | | Method: | SOLUTION NMR | | Cite: | Solution structure of Fe(II) cytochrome c551 from Pseudomonas aeruginosa as determined by two-dimensional 1H NMR.
Biochemistry, 30, 1991
|
|
1NKR
 
 | | INHIBITORY RECEPTOR (P58-CL42) FOR HUMAN NATURAL KILLER CELLS | | Descriptor: | P58-CL42 KIR | | Authors: | Fan, Q.R, Mosyak, L, Winter, C.C, Wagtmann, N, Long, E.O, Wiley, D.C. | | Deposit date: | 1998-06-24 | | Release date: | 1998-11-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of the inhibitory receptor for human natural killer cells resembles haematopoietic receptors.
Nature, 389, 1997
|
|
2G85
 
 | | Crystal structure of chorismate synthase from Mycobacterium tuberculosis at 2.22 angstrons of resolution | | Descriptor: | Chorismate synthase | | Authors: | Dias, M.V.B, dos Santos, B.B, Ely, F, Basso, L.A, Santos, D.S, de Azevedo Jr, W.F. | | Deposit date: | 2006-03-01 | | Release date: | 2007-03-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Crystal structure of chorismate synthase from Mycobacterium tuberculosis at 2.22 angstron of resolution
To be Published
|
|
2QIZ
 
 | |
2QR3
 
 | | Crystal structure of the N-terminal signal receiver domain of two-component system response regulator from Bacteroides fragilis | | Descriptor: | Two-component system response regulator | | Authors: | Bonanno, J.B, Freeman, J, Bain, K.T, Mendoza, M, Romero, R, Fong, R, Smith, D, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-07-27 | | Release date: | 2007-08-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the N-terminal signal receiver domain of two-component system response regulator from Bacteroides fragilis.
To be Published
|
|
1ZNF
 
 | | THREE-DIMENSIONAL SOLUTION STRUCTURE OF A SINGLE ZINC FINGER DNA-BINDING DOMAIN | | Descriptor: | 31ST ZINC FINGER FROM XFIN, ZINC ION | | Authors: | Lee, M.S, Gippert, G.P, Soman, K.V, Case, D.A, Wright, P.E. | | Deposit date: | 1989-09-25 | | Release date: | 1991-07-15 | | Last modified: | 2017-11-29 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional solution structure of a single zinc finger DNA-binding domain.
Science, 245, 1989
|
|
1YVG
 
 | | Structural analysis of the catalytic domain of tetanus neurotoxin | | Descriptor: | Tetanus toxin, light chain, ZINC ION | | Authors: | Rao, K.N, Kumaran, D, Binz, T, Swaminathan, S. | | Deposit date: | 2005-02-15 | | Release date: | 2005-03-22 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural analysis of the catalytic domain of tetanus neurotoxin.
Toxicon, 45, 2005
|
|
2ASE
 
 | |
2OVC
 
 | |
2GBH
 
 | | NMR structure of stem region of helix-35 of 23S E.coli ribosomal RNA (residues 736-760) | | Descriptor: | 5'-R(*(GMP)P*GP*GP*CP*UP*AP*AP*UP*GP*(PSU)P*UP*GP*AP*AP*AP*AP*AP*UP*UP*AP*GP*CP*CP*C)-3' | | Authors: | Bax, A, Boisbouvier, J, Bryce, D, Grishaev, A, Jaroniec, C, Miclet, E, Nikonovicz, E, O'Neil-Cabello, E, Ying, J. | | Deposit date: | 2006-03-10 | | Release date: | 2006-04-11 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Measurement of five dipolar couplings from a single 3D NMR multiplet applied to the study of RNA dynamics.
J.Am.Chem.Soc., 126, 2004
|
|
