2K74
 
 | | Solution NMR structure of DsbB-ubiquinone complex | | Descriptor: | Disulfide bond formation protein B, UBIQUINONE-2 | | Authors: | Zhou, Y, Cierpicki, T, Flores Jimenez, R.H, Lukasik, S.M, Ellena, J.F, Cafiso, D.S, Kadokura, H, Beckwith, J, Bushweller, J.H. | | Deposit date: | 2008-08-01 | | Release date: | 2008-10-07 | | Last modified: | 2024-11-27 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of the integral membrane enzyme DsbB: functional insights into DsbB-catalyzed disulfide bond formation.
Mol.Cell, 31, 2008
|
|
2K9B
 
 | | Structure and membrane interactions of the antibiotic peptide dermadistinctin K by multidimensional solution and oriented 15N and 31P solid-state NMR spectroscopy | | Descriptor: | Dermadistinctin-K | | Authors: | Moraes, C.M, Verly, R.M, Resende, J.M, Aisenbrey, C, Bemquerer, M.P, Pilo-Veloso, D, Valente, A, Almeida, F.C.L, Bechinger, B. | | Deposit date: | 2008-10-07 | | Release date: | 2009-04-14 | | Last modified: | 2024-11-20 | | Method: | SOLUTION NMR | | Cite: | Structure and membrane interactions of the antibiotic peptide dermadistinctin K by multidimensional solution and oriented 15N and 31P solid-state NMR spectroscopy.
Biophys.J., 96, 2009
|
|
6EPD
 
 | | Substrate processing state 26S proteasome (SPS1) | | Descriptor: | 26S proteasome non-ATPase regulatory subunit 1, 26S proteasome non-ATPase regulatory subunit 11, 26S proteasome non-ATPase regulatory subunit 13, ... | | Authors: | Guo, Q, Lehmer, C, Martinez-Sanchez, A, Rudack, T, Beck, F, Hartmann, H, Hipp, M.S, Hartl, F.U, Edbauer, D, Baumeister, W, Fernandez-Busnadiego, R. | | Deposit date: | 2017-10-11 | | Release date: | 2018-02-07 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (15.4 Å) | | Cite: | In Situ Structure of Neuronal C9orf72 Poly-GA Aggregates Reveals Proteasome Recruitment.
Cell, 172, 2018
|
|
2KGS
 
 | | Solution structure of the amino-terminal domain of OmpATb, a pore forming protein from Mycobacterium tuberculosis | | Descriptor: | Uncharacterized protein Rv0899/MT0922 | | Authors: | Yang, Y, Auguin, D, Delbecq, S, Dumas, E, Molle, V, Saint, N. | | Deposit date: | 2009-03-18 | | Release date: | 2010-03-09 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure of the Mycobacterium tuberculosis OmpATb protein: A model of an oligomeric channel in the mycobacterial cell wall
Proteins, 79, 2011
|
|
2KHW
 
 | | Solution Structure of the human Polymerase iota UBM2-Ubiquitin Complex | | Descriptor: | Immunoglobulin G-binding protein G, DNA polymerase iota, Ubiquitin | | Authors: | Bomar, M.G, D'Souza, S, Bienko, M, Dikic, I, Walker, G. | | Deposit date: | 2009-04-13 | | Release date: | 2010-02-23 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the human Polymerase iota UBM2-Ubiquitin Complex
To be Published
|
|
6ENU
 
 | | Polyproline-stalled ribosome in the presence of elongation-factor P (EF-P) | | Descriptor: | 16S ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Huter, P, Arenz, S, Wilson, D.N. | | Deposit date: | 2017-10-06 | | Release date: | 2017-11-22 | | Last modified: | 2025-04-09 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural Basis for Polyproline-Mediated Ribosome Stalling and Rescue by the Translation Elongation Factor EF-P.
Mol. Cell, 68, 2017
|
|
2KUK
 
 | |
5XFZ
 
 | | Crystal structure of a novel PET hydrolase R103G/S131A mutant from Ideonella sakaiensis 201-F6 | | Descriptor: | GLYCEROL, Poly(ethylene terephthalate) hydrolase, SULFATE ION | | Authors: | Han, X, Liu, W.D, Zheng, Y.Y, Chen, C.C, Guo, R.T. | | Deposit date: | 2017-04-11 | | Release date: | 2017-12-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural insight into catalytic mechanism of PET hydrolase
Nat Commun, 8, 2017
|
|
2KKV
 
 | | Solution NMR structure of an integrase domain from protein SPA4288 from Salmonella enterica, Northeast Structural Genomics Consortium Target SlR105H | | Descriptor: | Integrase | | Authors: | Mills, J.L, Wu, Y, Sukumaran, D.K, Belote, R.L, Ciccosanti, C, Jiang, M, Nair, R, Rost, B, Acton, T.B, Xiao, R, Swapna, G.V.T, Everett, J.K, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-06-29 | | Release date: | 2009-08-25 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of an integrase domain from protein SPA4288 from Salmonella enterica, Northeast Structural Genomics Consortium Target SlR105H
To be Published
|
|
5XH2
 
 | | Crystal structure of a novel PET hydrolase R103G/S131A mutant in complex with pNP from Ideonella sakaiensis 201-F6 | | Descriptor: | P-NITROPHENOL, Poly(ethylene terephthalate) hydrolase, SULFATE ION | | Authors: | Han, X, Liu, W.D, Zheng, Y.Y, Chen, C.C, Guo, R.T. | | Deposit date: | 2017-04-19 | | Release date: | 2017-12-20 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structural insight into catalytic mechanism of PET hydrolase
Nat Commun, 8, 2017
|
|
2FBO
 
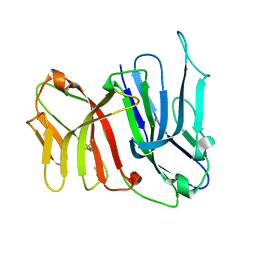 | | Crystal Structure of the Two Tandem V-type Regions of VCBP3 (v-region-containing chitin binding protein) to 1.85 A | | Descriptor: | variable region-containing chitin-binding protein 3 | | Authors: | Hernandez Prada, J.A, Haire, R.N, Jakoncic, J, Cannon, J.P, Litman, G.W, Ostrov, D.A. | | Deposit date: | 2005-12-09 | | Release date: | 2006-10-17 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Ancient evolutionary origin of diversified variable regions demonstrated by crystal structures of an immune-type receptor in amphioxus
Nat.Immunol., 7, 2006
|
|
2KXC
 
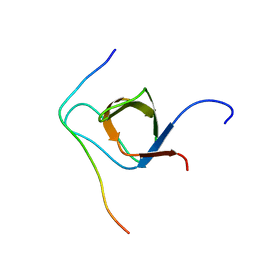 | | 1H, 13C, and 15N Chemical Shift Assignments for IRTKS-SH3 and EspFu-R47 complex | | Descriptor: | Brain-specific angiogenesis inhibitor 1-associated protein 2-like protein 1, EspF-like protein | | Authors: | Aitio, O, Hellman, M, Kazlauskas, A, Vingadassalom, D.F, Leong, J.M, Saksela, K, Permi, P. | | Deposit date: | 2010-04-30 | | Release date: | 2010-11-17 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Recognition of tandem PxxP motifs as a unique Src homology 3-binding mode triggers pathogen-driven actin assembly
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
2FCD
 
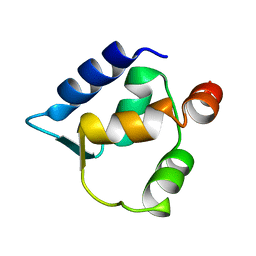 | | Solution structure of N-lobe Myosin Light Chain from Saccharomices cerevisiae | | Descriptor: | Myosin light chain 1 | | Authors: | Cicero, D.O, Pennestri, M, Contessa, G.M, Paci, M, Ragnini-Wilson, A, Melino, S. | | Deposit date: | 2005-12-12 | | Release date: | 2006-11-07 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structural basis for the interaction of the myosin light chain Mlc1p with the myosin V Myo2p IQ motifs.
J.Biol.Chem., 282, 2007
|
|
2KMZ
 
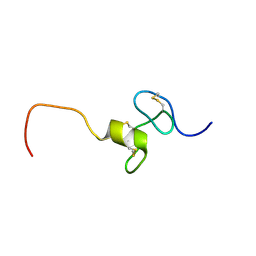 | | NMR Structure of hFn14 | | Descriptor: | Tumor necrosis factor receptor superfamily member 12A | | Authors: | Pellegrini, M, Willen, L, Perroud, M, Krushinskie, D, Strauch, K, Cuervo, H, Sun, Y, Day, E.S, Schneider, P, Zheng, T.S. | | Deposit date: | 2009-08-10 | | Release date: | 2011-06-29 | | Last modified: | 2024-11-20 | | Method: | SOLUTION NMR | | Cite: | Structure of the extracellular domains of human and Xenopus Fn14: implications in the evolution of TWEAK and Fn14 interactions.
Febs J., 280, 2013
|
|
6E9Y
 
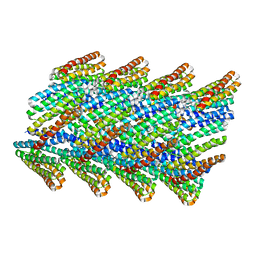 | | DHF38 filament | | Descriptor: | DHF38 filament | | Authors: | Lynch, E.M, Shen, H, Fallas, J.A, Kollman, J.M, Baker, D. | | Deposit date: | 2018-08-01 | | Release date: | 2018-11-21 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | De novo design of self-assembling helical protein filaments.
Science, 362, 2018
|
|
7N0E
 
 | |
6EC8
 
 | |
2KN5
 
 | |
6EDG
 
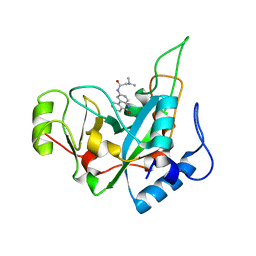 | | Pseudomonas exotoxin A domain III T18H477L | | Descriptor: | Exotoxin, N~2~,N~2~-DIMETHYL-N~1~-(6-OXO-5,6-DIHYDROPHENANTHRIDIN-2-YL)GLYCINAMIDE | | Authors: | Moss, D.L, Park, H.W, Mettu, R.R, Landry, S.J. | | Deposit date: | 2018-08-09 | | Release date: | 2019-02-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Deimmunizing substitutions in Pseudomonasexotoxin domain III perturb antigen processing without eliminating T-cell epitopes.
J.Biol.Chem., 294, 2019
|
|
1C8Q
 
 | |
2M21
 
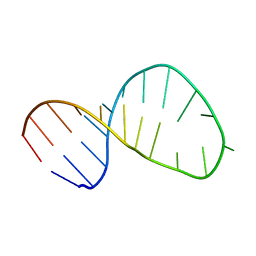 | | Solution structure of the Tetrahymena telomerase RNA stem IV terminal loop | | Descriptor: | 5'-R(*GP*GP*CP*GP*AP*UP*AP*CP*AP*CP*UP*AP*UP*UP*UP*AP*UP*CP*GP*CP*C)-3' | | Authors: | Cash, D.D, Richards, R.J, Wu, H, Trantirek, L, O'Connor, C.M, Feigon, J, Collins, K. | | Deposit date: | 2012-12-11 | | Release date: | 2013-03-20 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural study of elements of Tetrahymena telomerase RNA stem-loop IV domain important for function.
Rna, 12, 2006
|
|
2M24
 
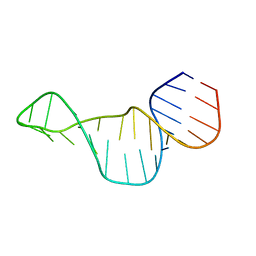 | |
7MRJ
 
 | | Crystal structure of a novel ubiquitin-like TINCR | | Descriptor: | ACETATE ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Forouhar, F, Morgado-Palacin, L, Brown, J.A, Martinez, T, Pedrero, J.M.G, Reglero, C, Chaudhry, I, Vaughan, J, Rodriguez-Perales, S, Allonca, E, Granda-Diaz, R, Quinn, S.A, Fernandez, A.F, Fraga, M.F, Kim, A.L, Santos-Juanes, J, Owens, D.M, Rodrigo, J.P, Saghatelian, A, Ferrando, A.A. | | Deposit date: | 2021-05-07 | | Release date: | 2022-03-16 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | The TINCR ubiquitin-like microprotein is a tumor suppressor in squamous cell carcinoma.
Nat Commun, 14, 2023
|
|
2MKP
 
 | | N domain of cardiac troponin C bound to the switch fragment of fast skeletal troponin I at pH 6 | | Descriptor: | CALCIUM ION, Troponin C, slow skeletal and cardiac muscles, ... | | Authors: | Robertson, I.M, Pineda-Sanabria, S.E, Holmes, P.C, Sykes, B.D. | | Deposit date: | 2014-02-11 | | Release date: | 2014-02-26 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Conformation of the critical pH sensitive region of troponin depends upon a single residue in troponin I.
Arch.Biochem.Biophys., 552-553, 2014
|
|
2MDV
 
 | |
