5GAD
 
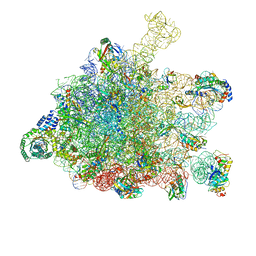 | | RNC-SRP-SR complex early state | | Descriptor: | 1A9L SS, 23S rRNA, 50S ribosomal protein L10, ... | | Authors: | Jomaa, A, Boehringer, D, Leibundgut, M, Ban, N. | | Deposit date: | 2015-11-24 | | Release date: | 2016-01-27 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structures of the E. coli translating ribosome with SRP and its receptor and with the translocon.
Nat Commun, 7, 2016
|
|
2MD0
 
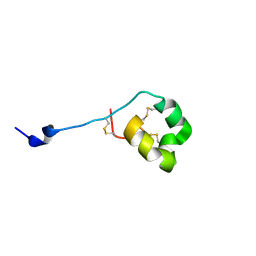 | | Solution structure of ShK-like immunomodulatory peptide from Ancylostoma caninum (hookworm) | | Descriptor: | AcK1 | | Authors: | Chhabra, S, Swarbrick, J.D, Mohanty, B, Chang, S.C, Chandy, G.K, Pennington, M.W, Norton, R.S. | | Deposit date: | 2013-08-28 | | Release date: | 2014-07-02 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Kv1.3 channel-blocking immunomodulatory peptides from parasitic worms: implications for autoimmune diseases.
Faseb J., 28, 2014
|
|
4PT5
 
 | |
2M8P
 
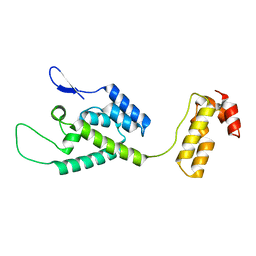 | | The structure of the W184AM185A mutant of the HIV-1 capsid protein | | Descriptor: | Capsid protein p24 | | Authors: | Deshmukh, L, Schwieters, C.D, Grishaev, A, Clore, G, Ghirlando, R. | | Deposit date: | 2013-05-24 | | Release date: | 2013-11-20 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR, SOLUTION SCATTERING | | Cite: | Structure and Dynamics of Full-Length HIV-1 Capsid Protein in Solution.
J.Am.Chem.Soc., 135, 2013
|
|
2M02
 
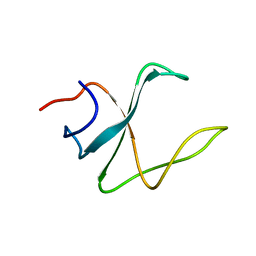 | | 3D structure of cap-gly domain of mammalian dynactin determined by magic angle spinning NMR spectroscopy | | Descriptor: | Dynactin subunit 1 | | Authors: | Yan, S, Hou, G, Schwieters, C.D, Ahmed, S, Williams, J.C, Polenova, T. | | Deposit date: | 2012-10-15 | | Release date: | 2013-05-08 | | Last modified: | 2024-05-15 | | Method: | SOLID-STATE NMR | | Cite: | Three-Dimensional Structure of CAP-Gly Domain of Mammalian Dynactin Determined by Magic Angle Spinning NMR Spectroscopy: Conformational Plasticity and Interactions with End-Binding Protein EB1.
J.Mol.Biol., 425, 2013
|
|
7JGW
 
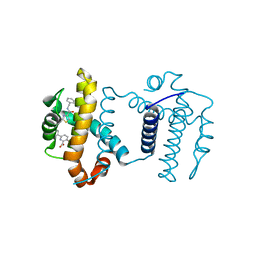 | | Crystal structure of BCL-XL in complex with COMPOUND 1620116, CRYSTAL FORM 1 | | Descriptor: | 6-[(8E)-8-{2-[4-(benzylcarbamoyl)-1,3-thiazol-2-yl]hydrazinylidene}-5,6,7,8-tetrahydronaphthalen-2-yl]-3-(2-phenylethoxy)pyridine-2-carboxylic acid, Bcl-2-like protein 1 | | Authors: | Lee, M, Fairlie, W.D, Smith, B.J, Lee, E.F. | | Deposit date: | 2020-07-19 | | Release date: | 2021-02-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Optimization of Benzothiazole and Thiazole Hydrazones as Inhibitors of Schistosome BCL-2.
Acs Infect Dis., 7, 2021
|
|
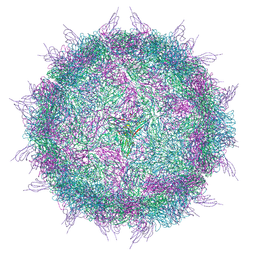
6EIT
 
 | |
1BRZ
 
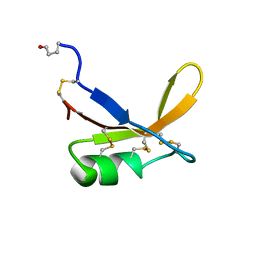 | | SOLUTION STRUCTURE OF THE SWEET PROTEIN BRAZZEIN, NMR, 43 STRUCTURES | | Descriptor: | BRAZZEIN | | Authors: | Caldwell, J.E, Abildgaard, F, Dzakula, Z, Ming, D, Hellekant, G, Markley, J.L. | | Deposit date: | 1998-03-12 | | Release date: | 1998-07-01 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the thermostable sweet-tasting protein brazzein.
Nat.Struct.Biol., 5, 1998
|
|
2M94
 
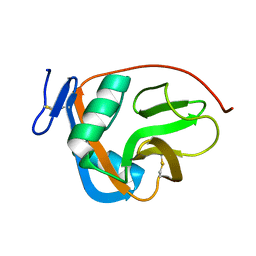 | |
2MAF
 
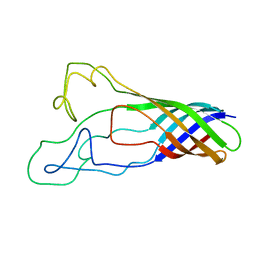 | | Solution structure of opa60 from n. gonorrhoeae | | Descriptor: | Opacity protein opA60 | | Authors: | Columbus, L, Fox, D.A, Larsson, P, Lo, R.H, Kroncke, B.M, Kasson, P.M. | | Deposit date: | 2013-07-08 | | Release date: | 2014-11-19 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure of the Neisserial outer membrane protein Opa60: loop flexibility essential to receptor recognition and bacterial engulfment.
J.Am.Chem.Soc., 136, 2014
|
|
2MC1
 
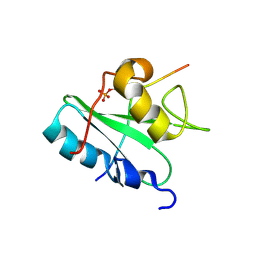 | | Solution structure of the Vav1 SH2 domain complexed with a Syk-derived singly phosphorylated peptide | | Descriptor: | Proto-oncogene vav, Tyrosine-protein kinase SYK | | Authors: | Chen, C, Piraner, D, Gorenstein, N.M, Geahlen, R.L, Post, C.B. | | Deposit date: | 2013-08-13 | | Release date: | 2013-08-28 | | Last modified: | 2024-11-20 | | Method: | SOLUTION NMR | | Cite: | Differential recognition of syk-binding sites by each of the two phosphotyrosine-binding pockets of the Vav SH2 domain.
Biopolymers, 99, 2013
|
|
2M4N
 
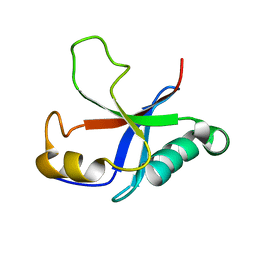 | | Solution structure of the putative Ras interaction domain of AFD-1, isoform a from Caenorhabditis elegans | | Descriptor: | Protein AFD-1, isoform a | | Authors: | Harris, R, Hillerich, B, Ahmed, M, Bonanno, J.B, Chamala, S, Evans, B, Lafleur, J, Hammonds, J, Washington, E, Stead, M, Love, J, Attonito, J, Seidel, R.D, Liddington, R.C, Weis, W.I, Nelson, W.J, Girvin, M.E, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC), Assembly, Dynamics and Evolution of Cell-Cell and Cell-Matrix Adhesions (CELLMAT) | | Deposit date: | 2013-02-07 | | Release date: | 2013-03-20 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the putative Ras interaction domain of AFD-1, isoform a from Caenorhabditis elegans
To be Published
|
|
7L33
 
 | | X-ray Structure of a Cu-Bound De Novo Designed Peptide Trimer | | Descriptor: | COPPER (II) ION, Cu-3SCC | | Authors: | Chakraborty, S, Wawrzak, Z, Prasad, P, Mitra, S, Prakash, D. | | Deposit date: | 2020-12-17 | | Release date: | 2021-08-11 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | De Novo Design of a Self-Assembled Artificial Copper Peptide that Activates and Reduces Peroxide
Acs Catalysis, 11, 2021
|
|
2MIF
 
 | |
3J32
 
 | | An asymmetric unit map from electron cryo-microscopy of Haliotis diversicolor molluscan hemocyanin isoform 1 (HdH1) | | Descriptor: | Hemocyanin isoform 1 | | Authors: | Zhang, Q, Dai, X, Cong, Y, Zhang, J, Chen, D.-H, Dougherty, M, Wang, J, Ludtke, S, Schmid, M.F, Chiu, W. | | Deposit date: | 2013-02-20 | | Release date: | 2013-04-17 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Cryo-EM structure of a molluscan hemocyanin suggests its allosteric mechanism.
Structure, 21, 2013
|
|
2LZ1
 
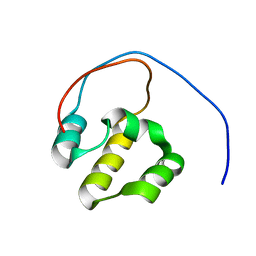 | | Solution NMR Structure of the DNA-Binding Domain of Human NF-E2-Related Factor 2, Northeast Structural Genomics Consortium (NESG) Target HR3520O | | Descriptor: | Nuclear factor erythroid 2-related factor 2 | | Authors: | Eletsky, A, Pulavarti, S.V.S.R.K, Lee, D, Kohan, E, Janjua, H, Xiao, R, Acton, T.B, Everett, J.K, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2012-09-22 | | Release date: | 2012-11-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Structure of the DNA-Binding Domain of Human NF-E2-Related Factor 2, Northeast Structural Genomics Consortium (NESG) Target HR3520O
To be Published
|
|
4PTI
 
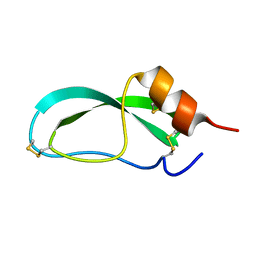 | | THE GEOMETRY OF THE REACTIVE SITE AND OF THE PEPTIDE GROUPS IN TRYPSIN, TRYPSINOGEN AND ITS COMPLEXES WITH INHIBITORS | | Descriptor: | TRYPSIN INHIBITOR | | Authors: | Huber, R, Kukla, D, Ruehlmann, A, Epp, O, Formanek, H, Deisenhofer, J, Steigemann, W. | | Deposit date: | 1982-09-27 | | Release date: | 1983-01-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The Geometry of the Reactive Site and of the Peptide Groups in Trypsin, Trypsinogen and its Complexes with Inhibitors
Acta Crystallogr.,Sect.B, 39, 1983
|
|
7L7J
 
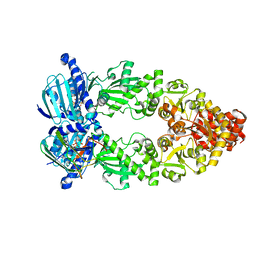 | | Cryo-EM structure of Hsp90:p23 closed-state complex | | Descriptor: | Heat shock protein HSP 90-alpha, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, Prostaglandin E synthase 3 | | Authors: | Lee, K, Thwin, A.C, Tse, E, Gates, S.N, Southworth, D.R. | | Deposit date: | 2020-12-28 | | Release date: | 2021-08-25 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | The structure of an Hsp90-immunophilin complex reveals cochaperone recognition of the client maturation state.
Mol.Cell, 81, 2021
|
|
2M47
 
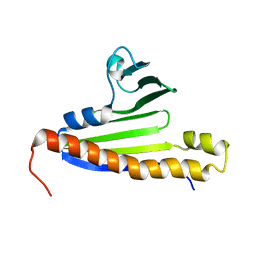 | | Solution NMR structure of the Polyketide_cyc-like protein Cgl2372 from Corynebacterium glutamicum, Northeast Structural Genomics Consortium Target CgR160 | | Descriptor: | Uncharacterized protein Cgl2373 | | Authors: | Yang, Y, Ramelot, T.A, Lee, D, Ciccosanti, C, Sapin, A, Janjua, H, Nair, R, Rost, B, Acton, T.B, Xiao, R, Everett, J.K, Montelione, G.T, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2013-01-30 | | Release date: | 2013-02-13 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of the Polyketide_cyc-like protein Cgl2372 from Corynebacterium glutamicum, Northeast Structural Genomics Consortium Target CgR160.
To be Published
|
|
7NAX
 
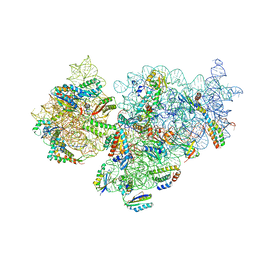 | | Complete Bacterial 30S ribosomal subunit assembly complex state I (Consensus Refinement) | | Descriptor: | 16S rRNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Schedlbauer, A, Iturrioz, I, Ochoa-Lizarralde, B, Diercks, T, Lopez-Alonso, J, Kaminishi, T, Capuni, R, Astigarraga, E, Gil-Carton, D, Fucini, P, Connell, S. | | Deposit date: | 2021-01-25 | | Release date: | 2021-12-08 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.96 Å) | | Cite: | A conserved rRNA switch is central to decoding site maturation on the small ribosomal subunit.
Sci Adv, 7, 2021
|
|
2M79
 
 | | [Asp2,11]RTD-1 | | Descriptor: | [Asp2,11]RTD-1 | | Authors: | Conibear, A.C, Bochen, A, Rosengren, K, Kessler, H, Craik, D.J. | | Deposit date: | 2013-04-19 | | Release date: | 2014-02-05 | | Last modified: | 2024-11-13 | | Method: | SOLUTION NMR | | Cite: | The Cyclic Cystine Ladder of Theta-Defensins as a Stable, Bifunctional Scaffold: A Proof-of-Concept Study Using the Integrin-Binding RGD Motif
Chembiochem, 15, 2014
|
|
2M8N
 
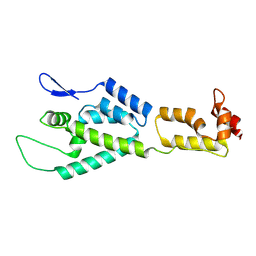 | | HIV-1 capsid monomer structure | | Descriptor: | Capsid protein p24 | | Authors: | Deshmukh, L, Schwieters, C.D, Grishaev, A, Clore, G, Ghirlando, R. | | Deposit date: | 2013-05-24 | | Release date: | 2013-11-20 | | Last modified: | 2024-11-27 | | Method: | SOLUTION NMR, SOLUTION SCATTERING | | Cite: | Structure and Dynamics of Full-Length HIV-1 Capsid Protein in Solution.
J.Am.Chem.Soc., 135, 2013
|
|
2LZH
 
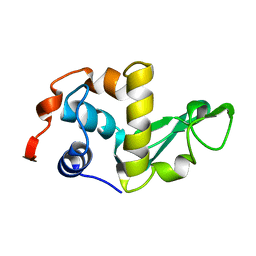 | | THE STRUCTURES OF THE MONOCLINIC AND ORTHORHOMBIC FORMS OF HEN EGG-WHITE LYSOZYME AT 6 ANGSTROMS RESOLUTION. | | Descriptor: | HEN EGG WHITE LYSOZYME | | Authors: | Artymiuk, P.J, Blake, C.C.F, Rice, D.W, Wilson, K.S. | | Deposit date: | 1981-06-29 | | Release date: | 1981-09-28 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (6 Å) | | Cite: | The Structures of the Monoclinic and Orthorhombic Forms of Hen Egg-White Lysozyme at 6 Angstroms Resolution.
Acta Crystallogr.,Sect.B, 38, 1982
|
|
7NAR
 
 | | Complete Bacterial 30S ribosomal subunit assembly complex state F (+RsgA)(Consensus Refinement) | | Descriptor: | 16S rRNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Schedlbauer, A, Iturrioz, I, Ochoa-Lizarralde, B, Diercks, T, Kaminishi, T, Capuni, R, Astigarraga, E, Gil-Carton, D, Fucini, P, Connell, S. | | Deposit date: | 2021-01-25 | | Release date: | 2021-12-08 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | A conserved rRNA switch is central to decoding site maturation on the small ribosomal subunit.
Sci Adv, 7, 2021
|
|
5GAG
 
 | | RNC in complex with SRP-SR in the closed state | | Descriptor: | 1A9L SS, 23S rRNA, 50S ribosomal protein L10, ... | | Authors: | Jomaa, A, Boehringer, D, Leibundgut, M, Ban, N. | | Deposit date: | 2015-11-26 | | Release date: | 2016-01-27 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structures of the E. coli translating ribosome with SRP and its receptor and with the translocon.
Nat Commun, 7, 2016
|
|
