5IBW
 
 | | Complex of MlcC bound to the tandem IQ motif of MyoC | | Descriptor: | Calcium-binding EF-hand domain-containing protein, Myosin IC heavy chain, SODIUM ION | | Authors: | Langelaan, D.N, Smith, S.P. | | Deposit date: | 2016-02-22 | | Release date: | 2016-08-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of the Single-lobe Myosin Light Chain C in Complex with the Light Chain-binding Domains of Myosin-1C Provides Insights into Divergent IQ Motif Recognition.
J.Biol.Chem., 291, 2016
|
|
7B4A
 
 | | Structural basis of reactivation of oncogenic p53 mutants by a small molecule: methylene quinuclidinone (MQ). Human p53DBD-R273H mutant bound to DNA: R273H-DNA | | Descriptor: | Cellular tumor antigen p53, DNA target, FORMIC ACID, ... | | Authors: | Golovenko, D, Rozenberg, H, Degtjarik, O, Shakked, Z. | | Deposit date: | 2020-12-02 | | Release date: | 2021-12-08 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis of reactivation of oncogenic p53 mutants by a small molecule: methylene quinuclidinone (MQ).
Nat Commun, 12, 2021
|
|
3FMG
 
 | | Structure of rotavirus outer capsid protein VP7 trimer in complex with a neutralizing Fab | | Descriptor: | CALCIUM ION, Fab of neutralizing antibody 4F8, heavy chain, ... | | Authors: | Aoki, S.T, Settembre, E.C, Trask, S.D, Greenberg, H.B, Harrison, S.C, Dormitzer, P.R. | | Deposit date: | 2008-12-22 | | Release date: | 2009-06-23 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structure of rotavirus outer-layer protein VP7 bound with a neutralizing Fab.
Science, 324, 2009
|
|
5FBX
 
 | | Crystal structure of the first bromodomain of human BRD4 in complex with PNZ5 isoxazole inhibitor | | Descriptor: | (3~{S})-5-(3,5-dimethyl-1,2-oxazol-4-yl)-2-methyl-3-phenyl-3~{H}-isoindol-1-one, Bromodomain-containing protein 4 | | Authors: | Tallant, C, Clark, P.G.K, Siejka, P, Fedorov, O, Nowak, R, Srikannathasan, V, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Muller, S, Brennan, P.E, Dixon, D, Knapp, S. | | Deposit date: | 2015-12-14 | | Release date: | 2015-12-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of the first bromodomain of human BRD4 in complex with PNZ5 isoxazole inhibitor
To Be Published
|
|
2JS1
 
 | | Solution NMR structure of the homodimer protein YVFG from Bacillus subtilis, Northeast Structural Genomics Consortium Target SR478 | | Descriptor: | Uncharacterized protein yvfG | | Authors: | Macnaughtan, M.A, Weldeghiorghis, T, Wang, X, Bansal, S, Tian, F, Wang, D, Janjua, H, Cunningham, K, Ma, L.-C, Xiao, R, Liu, J, Baran, M.C, Swapna, G.V.T, Acton, T.B, Rost, B, Montelione, G.T, Prestegard, J.H, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2007-06-29 | | Release date: | 2007-07-24 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | NMR Structure of the Bacillus subtilis Protein YvfG.
To be Published
|
|
3ILD
 
 | | Structure of ORF157-K57A from Acidianus filamentous virus 1 | | Descriptor: | MAGNESIUM ION, Putative uncharacterized protein | | Authors: | Goulet, A, Lichiere, J, Prangishvili, D, van Tilbeurgh, H, Cambillau, C, Campanacci, V. | | Deposit date: | 2009-08-07 | | Release date: | 2010-03-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | ORF157 from the archaeal virus Acidianus filamentous virus 1 defines a new class of nuclease
J.Virol., 84, 2010
|
|
6VWL
 
 | | 70S ribosome bound to HIV frameshifting stem-loop (FSS) and P/E tRNA (rotated conformation) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Loerch, S, Bao, C, Ling, C, Korostelev, A.A, Grigorieff, N, Ermolenko, D.M. | | Deposit date: | 2020-02-20 | | Release date: | 2020-06-03 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | mRNA stem-loops can pause the ribosome by hindering A-site tRNA binding.
Elife, 9, 2020
|
|
7FU6
 
 | | Crystal Structure of human cyclic GMP-AMP synthase in complex with 2-benzyl-6-(2-chloro-4-methylphenyl)indazole-4-carboxylic acid | | Descriptor: | (6M)-2-benzyl-6-(2-chloro-4-methylphenyl)-2H-indazole-4-carboxylic acid, Cyclic GMP-AMP synthase, ZINC ION | | Authors: | Leibrock, L, Benz, J, Groebke-Zbinden, K, Hunziker, D, Rudolph, M.G. | | Deposit date: | 2023-02-08 | | Release date: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Crystal Structure of a human cyclic GMP-AMP synthase complex
To be published
|
|
2JZ6
 
 | | Solution structure of 50S ribosomal protein L28 from Thermotoga maritima. Northeast Structural Genomics Consortium target VR97 | | Descriptor: | 50S ribosomal protein L28 | | Authors: | Wu, Y, Singarapu, K.K, Guido, V, Yee, A, Sukumaran, D, Arrowsmith, C.H, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2007-12-28 | | Release date: | 2008-01-29 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Solution structure of 50S ribosomal protein L28 from Thermotoga maritima. Northeast Structural Genomics Consortium target VR97.
To be Published
|
|
2JQ1
 
 | | Phylloseptin-3 | | Descriptor: | Phylloseptin-3 | | Authors: | Resende, J.M, Mendonca Moraes, C, Almeida, F.C.L, Prates, M.V, Cesar, A, Valente, A, Bemquerer, M.P, Pilo-Veloso, D, Bechinger, B. | | Deposit date: | 2007-05-25 | | Release date: | 2008-06-03 | | Last modified: | 2024-11-20 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structures of the antimicrobial peptides phylloseptin-1, -2, and -3 and biological activity: The role of charges and hydrogen bonding interactions in stabilizing helix conformations
Peptides, 29, 2008
|
|
8CRD
 
 | |
2JN3
 
 | | NMR structure of cl-BABP complexed to chenodeoxycholic acid | | Descriptor: | CHENODEOXYCHOLIC ACID, Fatty acid-binding protein, liver | | Authors: | Eliseo, T, Ragona, L, Catalano, M, Assfalf, M, Paci, M, Zetta, L, Molinari, H, Cicero, D.O. | | Deposit date: | 2006-12-22 | | Release date: | 2007-07-03 | | Last modified: | 2023-12-20 | | Method: | SOLUTION NMR | | Cite: | Structural and dynamic determinants of ligand binding in the ternary complex of chicken liver bile acid binding protein with two bile salts revealed by NMR
Biochemistry, 46, 2007
|
|
2JNR
 
 | | Discovery and optimization of a natural HIV-1 entry inhibitor targeting the gp41 fusion peptide | | Descriptor: | ENV polyprotein, VIR165 | | Authors: | Munch, J, Standker, L, Adermann, K, Schulz, A, Pohlmann, S, Chaipan, C, Biet, T, Peters, T, Meyer, B, Wilhelm, D, Lu, H, Jing, W, Jiang, S, Forssmann, W, Kirchhoff, F. | | Deposit date: | 2007-02-01 | | Release date: | 2007-05-08 | | Last modified: | 2024-11-20 | | Method: | SOLUTION NMR | | Cite: | Discovery and Optimization of a Natural HIV-1 Entry Inhibitor Targeting the gp41 Fusion Peptide.
Cell(Cambridge,Mass.), 129, 2007
|
|
2JQT
 
 | | Structure of the bacterial replication origin-associated protein Cnu | | Descriptor: | H-NS/stpA-binding protein 2 | | Authors: | Bae, S.H, Liu, D, Lim, H.M, Lee, Y, Choi, B.S. | | Deposit date: | 2007-06-07 | | Release date: | 2008-04-22 | | Last modified: | 2023-12-20 | | Method: | SOLUTION NMR | | Cite: | Structure of the nucleoid-associated protein Cnu reveals common binding sites for H-NS in Cnu and Hha.
Biochemistry, 47, 2008
|
|
3F3V
 
 | | Kinase domain of cSrc in complex with inhibitor RL45 (Type II) | | Descriptor: | 1-{4-[(6-aminoquinazolin-4-yl)amino]phenyl}-3-[3-tert-butyl-1-(3-methylphenyl)-1H-pyrazol-5-yl]urea, Proto-oncogene tyrosine-protein kinase Src | | Authors: | Grutter, C, Kluter, S, Getlik, M, Rauh, D. | | Deposit date: | 2008-10-31 | | Release date: | 2009-06-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Hybrid compound design to overcome the gatekeeper T338M mutation in cSrc
J.Med.Chem., 52, 2009
|
|
7B4M
 
 | |
2JRQ
 
 | | NMR solution structure of the anticodon of E. coli TRNA-VAL3 with 1 modification (cmo5U34) | | Descriptor: | 5'-R(*CP*CP*UP*CP*CP*CP*UP*(CM0)P*AP*CP*AP*AP*GP*GP*AP*GP*G)-3' | | Authors: | Vendeix, F.A.P, Dziergowska, A, Gustilo, E.M, Graham, W.D, Sproat, B, Malkiewicz, A, Agris, P.F. | | Deposit date: | 2007-06-28 | | Release date: | 2007-07-24 | | Last modified: | 2023-12-20 | | Method: | SOLUTION NMR | | Cite: | Wobble-Position Modifications Pre-structure tRNA's Anticodon for Ribosome-Mediated Codon Binding
To be Published
|
|
7GAN
 
 | | PanDDA analysis group deposition -- Crystal Structure of MAP1LC3B in complex with Z56767614 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Microtubule-associated proteins 1A/1B light chain 3B, ... | | Authors: | Kumar, A, Marples, P.G, Tomlinson, C.W.E, Fearon, D, von-Delft, F, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-08-10 | | Release date: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
7B6W
 
 | | Crystal structure of the human alpha1B adrenergic receptor in complex with inverse agonist (+)-cyclazosin | | Descriptor: | Alpha-1B adrenergic receptor,alpha1B adrenergic receptor,Alpha-1B adrenergic receptor,alpha1B adrenergic receptor,Alpha-1B adrenergic receptor,alpha1B adrenergic receptor,Alpha-1B adrenergic receptor,alpha1B adrenergic receptor, [(4~{a}~{R},8~{a}~{S})-4-(4-azanyl-6,7-dimethoxy-quinazolin-2-yl)-2,3,4~{a},5,6,7,8,8~{a}-octahydroquinoxalin-1-yl]-(furan-2-yl)methanone | | Authors: | Deluigi, M, Morstein, L, Hilge, M, Schuster, M, Merklinger, L, Klipp, A, Scott, D.J, Plueckthun, A. | | Deposit date: | 2020-12-08 | | Release date: | 2022-01-12 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.873 Å) | | Cite: | Crystal structure of the alpha 1B -adrenergic receptor reveals molecular determinants of selective ligand recognition.
Nat Commun, 13, 2022
|
|
5IKI
 
 | | CYP106A2 WITH SUBSTRATE ABIETIC ACID | | Descriptor: | Abietic acid, Cytochrome P450(MEG), PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Janocha, S, Carius, Y, Bernhardt, R, Lancaster, C.R.D. | | Deposit date: | 2016-03-03 | | Release date: | 2016-05-25 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of CYP106A2 in Substrate-Free and Substrate-Bound Form.
Chembiochem, 17, 2016
|
|
7GAB
 
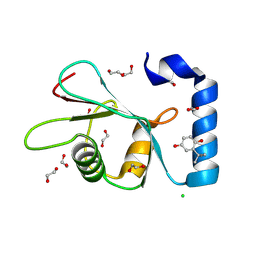 | | PanDDA analysis group deposition -- Crystal Structure of MAP1LC3B in complex with Z1255402624 | | Descriptor: | 1,2-ETHANEDIOL, 2-tert-butylbenzene-1,4-diol, CHLORIDE ION, ... | | Authors: | Kumar, A, Marples, P.G, Tomlinson, C.W.E, Fearon, D, von-Delft, F, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-08-10 | | Release date: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
7GAM
 
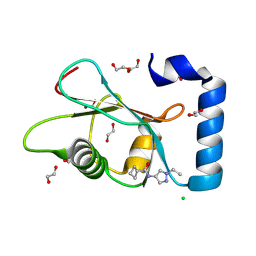 | | PanDDA analysis group deposition -- Crystal Structure of MAP1LC3B in complex with Z373768900 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Microtubule-associated proteins 1A/1B light chain 3B, ... | | Authors: | Kumar, A, Marples, P.G, Tomlinson, C.W.E, Fearon, D, von-Delft, F, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-08-10 | | Release date: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
2JMX
 
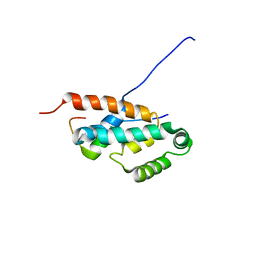 | | OSCP-NT (1-120) in complex with N-terminal (1-25) alpha subunit from F1-ATPase | | Descriptor: | ATP synthase O subunit, mitochondrial, ATP synthase subunit alpha heart isoform | | Authors: | Carbajo, R.J, Neuhaus, D, Kellas, F.A, Yang, J, Runswick, M.J, Montgomery, M.G, Walker, J.E. | | Deposit date: | 2006-12-12 | | Release date: | 2007-04-24 | | Last modified: | 2023-12-20 | | Method: | SOLUTION NMR | | Cite: | How the N-terminal Domain of the OSCP Subunit of Bovine F(1)F(o)-ATP Synthase Interacts with the N-terminal Region of an Alpha Subunit
J.Mol.Biol., 368, 2007
|
|
7GAQ
 
 | | PanDDA analysis group deposition -- Crystal Structure of MAP1LC3B in complex with Z755044716 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Microtubule-associated proteins 1A/1B light chain 3B, ... | | Authors: | Kumar, A, Marples, P.G, Tomlinson, C.W.E, Fearon, D, von-Delft, F, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-08-10 | | Release date: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
7GAL
 
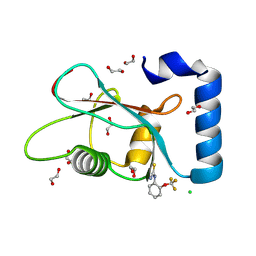 | | PanDDA analysis group deposition -- Crystal Structure of MAP1LC3B in complex with Z291279160 | | Descriptor: | 1,2-ETHANEDIOL, 1-[2-(trifluoromethyloxy)phenyl]thiourea, CHLORIDE ION, ... | | Authors: | Kumar, A, Marples, P.G, Tomlinson, C.W.E, Fearon, D, von-Delft, F, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-08-10 | | Release date: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
