8KDB
 
 | | Cryo-EM structure of the human parainfluenza virus hPIV3 L-P polymerase in dimeric form | | Descriptor: | MAGNESIUM ION, Phosphoprotein, RNA-directed RNA polymerase L, ... | | Authors: | Xie, J, Wang, L, Zhai, G, Wu, D, Lin, Z, Wang, M, Yan, X, Gao, L, Huang, X, Fearns, R, Chen, S. | | Deposit date: | 2023-08-09 | | Release date: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structural basis for dimerization of a paramyxovirus polymerase complex.
Nat Commun, 15, 2024
|
|
2F2J
 
 | |
7JR9
 
 | | Chlamydomonas reinhardtii radial spoke minimal head complex | | Descriptor: | Flagellar radial spoke protein 10, Flagellar radial spoke protein 4, Flagellar radial spoke protein 6, ... | | Authors: | Grossman-Haham, I, Coudray, N, Yu, Z, Wang, F, Bhabha, G, Vale, R.D. | | Deposit date: | 2020-08-11 | | Release date: | 2020-12-16 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.95 Å) | | Cite: | Structure of the radial spoke head and insights into its role in mechanoregulation of ciliary beating.
Nat.Struct.Mol.Biol., 28, 2021
|
|
9E2M
 
 | |
9E2L
 
 | |
9DTD
 
 | |
8TTE
 
 | | Protonated state of NorA at pH 5.0 | | Descriptor: | FabDA1 CDRH3 loop, Quinolone resistance protein NorA | | Authors: | Li, J.P, Li, Y, Koide, A, Kuang, H.H, Torres, V.J, Koide, S, Wang, D.N, Traaseth, N.J. | | Deposit date: | 2023-08-13 | | Release date: | 2024-05-29 | | Last modified: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (3.26 Å) | | Cite: | Proton-coupled transport mechanism of the efflux pump NorA.
Nat Commun, 15, 2024
|
|
6ZY0
 
 | | Catabolic reductive dehalogenase NpRdhA, N-terminally tagged, K488Q variant | | Descriptor: | CHLORIDE ION, COBALAMIN, IRON/SULFUR CLUSTER, ... | | Authors: | Leys, D, Halliwell, T. | | Deposit date: | 2020-07-30 | | Release date: | 2020-09-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Catabolic Reductive Dehalogenase Substrate Complex Structures Underpin Rational Repurposing of Substrate Scope.
Microorganisms, 8, 2020
|
|
9EJY
 
 | | Human hnRNPR extended eRRM1 domain | | Descriptor: | Heterogeneous nuclear ribonucleoprotein R | | Authors: | Eichhorn, C.D, Atsrim, E.S. | | Deposit date: | 2024-11-29 | | Release date: | 2025-04-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | An evolutionarily conserved tryptophan cage promotes folding of the extended RNA recognition motif in the hnRNPR-like protein family.
Protein Sci., 34, 2025
|
|
2J8J
 
 | | Solution Structure of the A4 Domain of Blood Coagulation Factor XI | | Descriptor: | COAGULATION FACTOR XI | | Authors: | Samuel, D, Cheng, H, Riley, P.W, Canutescu, A.A, Bu, Z, Walsh, P.N, Roder, H. | | Deposit date: | 2006-10-25 | | Release date: | 2007-10-02 | | Last modified: | 2024-11-13 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the A4 Domain of Factor Xi Sheds Light on the Mechanism of Zymogen Activation.
Proc.Natl.Acad.Sci.USA, 104, 2007
|
|
5FWK
 
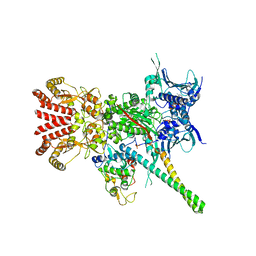 | | Atomic cryoEM structure of Hsp90-Cdc37-Cdk4 complex | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CYCLIN-DEPENDENT KINASE 4, HEAT SHOCK PROTEIN HSP 90 BETA, ... | | Authors: | Verba, K.A, Wang, R.Y.R, Arakawa, A, Liu, Y, Yokoyama, S, Agard, D.A. | | Deposit date: | 2016-02-17 | | Release date: | 2016-07-06 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Atomic Structure of Hsp90-Cdc37-Cdk4 Reveals that Hsp90 Traps and Stabilizes an Unfolded Kinase.
Science, 352, 2016
|
|
8TTG
 
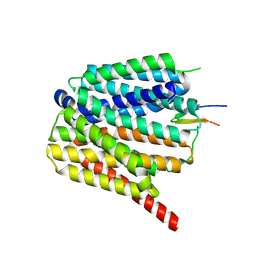 | | NorA single mutant - E222Q at pH 7.5 | | Descriptor: | FabDA1 CDRH3 loop, Quinolone resistance protein NorA | | Authors: | Li, J.P, Li, Y, Koide, A, Kuang, H.H, Torres, V.J, Koide, S, Wang, D.N, Traaseth, N.J. | | Deposit date: | 2023-08-13 | | Release date: | 2024-05-29 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (3.35 Å) | | Cite: | Proton-coupled transport mechanism of the efflux pump NorA.
Nat Commun, 15, 2024
|
|
8QZF
 
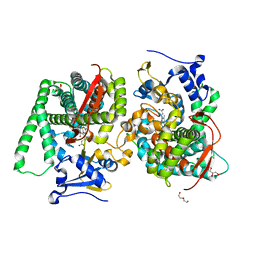 | | Heme-domain BM3 mutant T268E | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Bifunctional cytochrome P450/NADPH--P450 reductase, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Opperman, D.J, Ebrecht, A.C, Aschenbrenner, J.C. | | Deposit date: | 2023-10-27 | | Release date: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Revisiting strategies and their combinatorial effect for introducing peroxygenase activity in CYP102A1 (P450BM3)
Mol Catal, 557, 2024
|
|
8QZZ
 
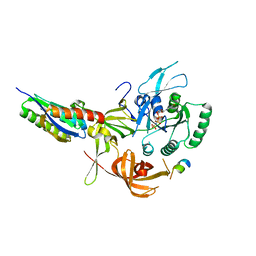 | | Crystal structure of human eIF2 alpha-gamma complexed with PPP1R15A_420-452 | | Descriptor: | Eukaryotic translation initiation factor 2 subunit 1, Eukaryotic translation initiation factor 2 subunit 3, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, ... | | Authors: | Yan, Y, Ron, D. | | Deposit date: | 2023-10-30 | | Release date: | 2024-03-20 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | Substrate recruitment via eIF2 gamma enhances catalytic efficiency of a holophosphatase that terminates the integrated stress response.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
7DZW
 
 | | Apo spike protein from SARS-CoV2 | | Descriptor: | Spike glycoprotein | | Authors: | Liu, Y, Soh, W.T, Li, S, Kishikawa, J, Hirose, M, Kato, T, Standley, D, Okada, M, Arase, H. | | Deposit date: | 2021-01-26 | | Release date: | 2021-06-02 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.45 Å) | | Cite: | An infectivity-enhancing site on the SARS-CoV-2 spike protein targeted by antibodies.
Cell, 184, 2021
|
|
8KDC
 
 | | Cryo-EM structure of the human parainfluenza virus hPIV3 L-P polymerase in monomeric form | | Descriptor: | MAGNESIUM ION, Phosphoprotein, RNA-directed RNA polymerase L, ... | | Authors: | Xie, J, Wang, L, Zhai, G, Wu, D, Lin, Z, Wang, M, Yan, X, Gao, L, Huang, X, Fearns, R, Chen, S. | | Deposit date: | 2023-08-09 | | Release date: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis for dimerization of a paramyxovirus polymerase complex.
Nat Commun, 15, 2024
|
|
5FET
 
 | | Crystal Structure of PVX_084705 in presence of Compound 2 | | Descriptor: | 4-[7-[(dimethylamino)methyl]-2-(4-fluorophenyl)imidazo[1,2-a]pyridin-3-yl]pyrimidin-2-amine, cGMP-dependent protein kinase, putative | | Authors: | Wernimont, A.K, Tempel, W, Walker, J.R, He, H, Seitova, A, Hills, T, Neculai, A.M, Baker, D.A, Flueck, C, Kettleborough, C.A, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Hui, R, Hutchinson, A, El Bakkouri, M, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-12-17 | | Release date: | 2016-01-13 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3.07 Å) | | Cite: | Crystal Structure of PVX_084705 in presence of Compound 2
To be published
|
|
7MKY
 
 | | SARS-CoV-2 frameshifting pseudoknot RNA | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, COBALT HEXAMMINE(III), MAGNESIUM ION, ... | | Authors: | Jones, C.P, Ferre-D'Amare, A.R. | | Deposit date: | 2021-04-27 | | Release date: | 2021-05-26 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | Crystal structure of the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) frameshifting pseudoknot.
Rna, 28, 2022
|
|
6ZT1
 
 | |
7JRJ
 
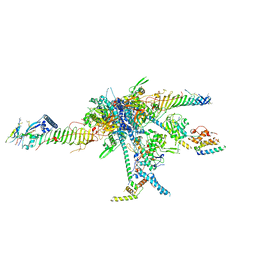 | | Chlamydomonas reinhardtii radial spoke head and neck (recombinant) | | Descriptor: | Flagellar radial spoke protein 1, Flagellar radial spoke protein 10, Flagellar radial spoke protein 2, ... | | Authors: | Grossman-Haham, I, Coudray, N, Yu, Z, Wang, F, Zhang, N, Bhabha, G, Vale, R.D. | | Deposit date: | 2020-08-12 | | Release date: | 2020-12-16 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.03 Å) | | Cite: | Structure of the radial spoke head and insights into its role in mechanoregulation of ciliary beating.
Nat.Struct.Mol.Biol., 28, 2021
|
|
3I5M
 
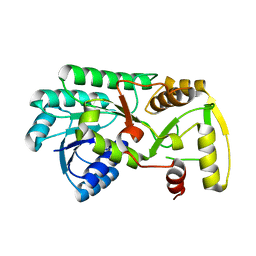 | | Structure of the apo form of leucoanthocyanidin reductase from vitis vinifera | | Descriptor: | Putative leucoanthocyanidin reductase 1 | | Authors: | Mauge, C, Gargouri, M, d'Estaintot, B.L, Granier, T, Gallois, B. | | Deposit date: | 2009-07-06 | | Release date: | 2010-02-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | Crystal structure and catalytic mechanism of leucoanthocyanidin reductase from Vitis vinifera.
J.Mol.Biol., 397, 2010
|
|
6DH9
 
 | |
8T3N
 
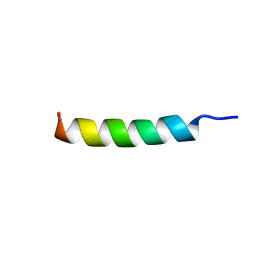 | | Solution NMR structure of synthetic peptide AMPCry10Aa_5 rational designed from Cry10Aa bacterial protein | | Descriptor: | Pesticidal crystal protein Cry10Aa peptide | | Authors: | Barra, J.B, Freitas, C.D.P, Rios, T.B, Maximiano, M.R, Fernandes, F.C, Amorim, G.C, Porto, W.F, Grossi-de-Sa, M.F, Franco, O.F, Liao, L.M. | | Deposit date: | 2023-06-07 | | Release date: | 2024-06-12 | | Last modified: | 2024-11-13 | | Method: | SOLUTION NMR | | Cite: | Anti-Staphy Peptides Rationally Designed from Cry10Aa Bacterial Protein.
Acs Omega, 9, 2024
|
|
9E2K
 
 | |
8EQ1
 
 | |
