8D6G
 
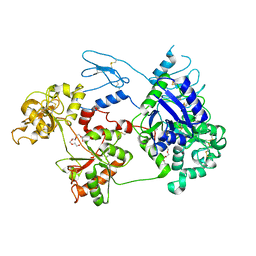 | | Nanorana parkeri saxiphilin | | Descriptor: | PENTAETHYLENE GLYCOL, Saxiphilin | | Authors: | Zakrzewska, S, Chen, Z, Minor, D.L. | | Deposit date: | 2022-06-06 | | Release date: | 2022-11-02 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Definition of a saxitoxin (STX) binding code enables discovery and characterization of the anuran saxiphilin family.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
8D6O
 
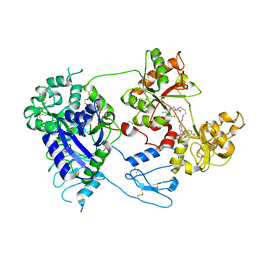 | | Nanorana parkeri saxiphilin:F-STX (soaked) | | Descriptor: | (2P)-4-({6-[({[(3aS,4R,7R,10aS)-2,6-diamino-10,10-dihydroxy-3a,4,9,10-tetrahydro-3H,8H-pyrrolo[1,2-c]purin-4-yl]methoxy}carbonyl)amino]hexyl}carbamoyl)-2-{[4aP,9(9a)P]-6-hydroxy-3-oxo-3H-xanthen-9-yl}benzoic acid, PENTAETHYLENE GLYCOL, Saxiphilin | | Authors: | Zakrzewska, S, Chen, Z, Minor, D.L. | | Deposit date: | 2022-06-06 | | Release date: | 2022-11-02 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Definition of a saxitoxin (STX) binding code enables discovery and characterization of the anuran saxiphilin family.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
8DTL
 
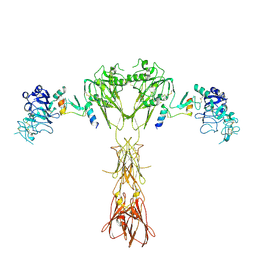 | | Cryo-EM structure of insulin receptor (IR) bound with S597 peptide | | Descriptor: | Insulin mimetic peptide S597, Insulin receptor | | Authors: | Park, J, Li, J, Mayer, J.P, Ball, K.A, Wu, J.Y, Hall, C, Accili, D, Stowell, M.H.B, Bai, X.C, Choi, E. | | Deposit date: | 2022-07-25 | | Release date: | 2022-09-07 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (5.4 Å) | | Cite: | Activation of the insulin receptor by an insulin mimetic peptide.
Nat Commun, 13, 2022
|
|
8D3M
 
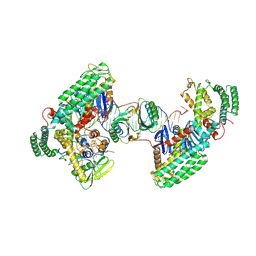 | | Type I-C Cas4-Cas1-Cas2 complex bound to a PAM/Processed prespacer | | Descriptor: | CRISPR-associated endonuclease Cas1, CRISPR-associated endonuclease Cas2, CRISPR-associated exonuclease Cas4, ... | | Authors: | Dhingra, Y, Suresh, S.K, Juneja, P, Sashital, D.G. | | Deposit date: | 2022-06-01 | | Release date: | 2022-11-02 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (3.41 Å) | | Cite: | PAM binding ensures orientational integration during Cas4-Cas1-Cas2-mediated CRISPR adaptation.
Mol.Cell, 82, 2022
|
|
7OF8
 
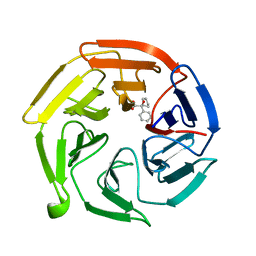 | | Keap1 kelch domain bound to a small molecule fragment | | Descriptor: | (2~{S})-2-cyclopentyl-2-oxidanyl-2-phenyl-ethanoic acid, Kelch-like ECH-associated protein 1 | | Authors: | Narayanan, D, Bach, A, Gajhede, M. | | Deposit date: | 2021-05-04 | | Release date: | 2022-11-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Development of Noncovalent Small-Molecule Keap1-Nrf2 Inhibitors by Fragment-Based Drug Discovery.
J.Med.Chem., 65, 2022
|
|
8D6P
 
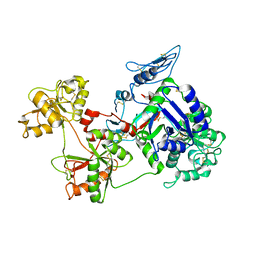 | |
2MAI
 
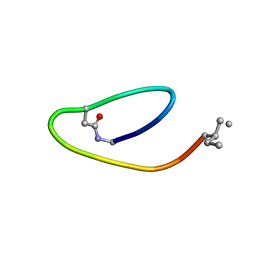 | | NMR structure of lassomycin | | Descriptor: | Lassomycin | | Authors: | Gavrish, E, Sit, C.S, Kandror, O, Spoering, A, Peoples, A, Ling, L, Fetterman, A, Hughes, D, Cao, S, Bissell, A, Torrey, H, Akopian, T, Mueller, A, Epstein, S, Goldberg, A, Clardy, J, Lewis, K. | | Deposit date: | 2013-07-09 | | Release date: | 2014-05-07 | | Last modified: | 2023-11-15 | | Method: | SOLUTION NMR | | Cite: | Lassomycin, a Ribosomally Synthesized Cyclic Peptide, Kills Mycobacterium tuberculosis by Targeting the ATP-Dependent Protease ClpC1P1P2.
Chem.Biol., 21, 2014
|
|
7BZJ
 
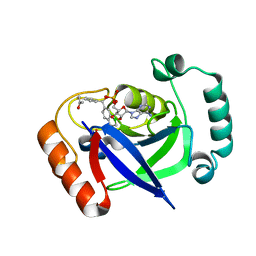 | | The Discovery of Benzhydrol-Oxaborole Hybrid Derivatives as Leucyl-tRNA Synthetase Inhibitors | | Descriptor: | Leucine--tRNA ligase, [(1~{R},5~{R},6~{S},8~{R})-8-(6-aminopurin-9-yl)-4'-[(~{R})-oxidanyl-[4-(2-oxidanylidenepropylsulfanyl)phenyl]methyl]spiro[2,4,7-trioxa-3-boranuidabicyclo[3.3.0]octane-3,7'-7-boranuidabicyclo[4.3.0]nona-1(6),2,4-triene]-6-yl]methoxy-tris(oxidanyl)phosphanium | | Authors: | Liu, R.J, Li, H, Wang, E.D, Zhou, H. | | Deposit date: | 2020-04-28 | | Release date: | 2020-12-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of benzhydrol-oxaborole derivatives as Streptococcus pneumoniae leucyl-tRNA synthetase inhibitors.
Bioorg.Med.Chem., 29, 2021
|
|
6VLE
 
 | | Crystal structure of human alpha 1,6-fucosyltransferase, FUT8 in its Apo-form | | Descriptor: | Alpha-(1,6)-fucosyltransferase, SULFATE ION | | Authors: | Jarva, M.A, Dramicanin, M, Lingford, J.P, Mao, R, John, A, Goddard-Borger, E.D. | | Deposit date: | 2020-01-23 | | Release date: | 2020-02-26 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Structural basis of substrate recognition and catalysis by fucosyltransferase 8.
J.Biol.Chem., 295, 2020
|
|
8D3L
 
 | | Type I-C Cas4-Cas1-Cas2 complex bound to a PAM/PAM prespacer | | Descriptor: | CRISPR-associated endonuclease Cas1, CRISPR-associated endonuclease Cas2, CRISPR-associated exonuclease Cas4, ... | | Authors: | Dhingra, Y, Suresh, S.K, Juneja, P, Sashital, D.G. | | Deposit date: | 2022-06-01 | | Release date: | 2022-11-02 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (3.49 Å) | | Cite: | PAM binding ensures orientational integration during Cas4-Cas1-Cas2-mediated CRISPR adaptation.
Mol.Cell, 82, 2022
|
|
8D6Q
 
 | |
2LXB
 
 | |
8DOY
 
 | | Crystal structure of SARS-CoV-2 main protease in complex with an inhibitor TKB-198 | | Descriptor: | 2-(2-METHOXYETHOXY)ETHANOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 3C-like proteinase nsp5, ... | | Authors: | Bulut, H, Hayashi, H, Tsuji, K, Kuwata, N, Das, D, Tamamura, H, Mitsuya, H. | | Deposit date: | 2022-07-14 | | Release date: | 2022-08-24 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Potent and biostable inhibitors of the main protease of SARS-CoV-2.
Iscience, 25, 2022
|
|
8D6S
 
 | | Rana catesbeiana saxiphilin mutant - Y558A:STX (co-crystal) | | Descriptor: | Saxiphilin, [(3aS,4R,10aS)-2,6-diamino-10,10-dihydroxy-3a,4,9,10-tetrahydro-3H,8H-pyrrolo[1,2-c]purin-4-yl]methyl carbamate | | Authors: | Chen, Z, Zakrzewska, S, Minor, D.L. | | Deposit date: | 2022-06-06 | | Release date: | 2022-11-02 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Definition of a saxitoxin (STX) binding code enables discovery and characterization of the anuran saxiphilin family.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
2MN1
 
 | | Solution Structure of kalata B1[W23WW] | | Descriptor: | kalata B1[W23WW] | | Authors: | Henriques, S.T, Huang, Y.H, Chaousis, S, Wang, C.K, Craik, D.J. | | Deposit date: | 2014-03-26 | | Release date: | 2015-05-27 | | Last modified: | 2024-11-20 | | Method: | SOLUTION NMR | | Cite: | Anticancer and toxic properties of cyclotides are dependent on phosphatidylethanolamine phospholipid targeting.
Chembiochem, 15, 2014
|
|
8E1Y
 
 | | Crystal Structure of SARS-CoV-2 Main protease A193S mutant in complex with Nirmatrelvir | | Descriptor: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5, DIMETHYL SULFOXIDE | | Authors: | Noske, G.D, Oliva, G, Godoy, A.S. | | Deposit date: | 2022-08-11 | | Release date: | 2022-08-24 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Structural basis of nirmatrelvir and ensitrelvir activity against naturally occurring polymorphisms of the SARS-CoV-2 main protease.
J.Biol.Chem., 299, 2023
|
|
2M6A
 
 | | NMR spatial structure of the antimicrobial peptide Tk-Amp-X2 | | Descriptor: | Predicted protein | | Authors: | Usmanova, D.R, Mineev, K.S, Arseniev, A.S, Berkut, A.A, Oparin, P.B, Grishin, E.V, Vassilevski, A.A. | | Deposit date: | 2013-03-28 | | Release date: | 2014-04-02 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Structural similarity between defense peptide from wheat and scorpion neurotoxin permits rational functional design
J.Biol.Chem., 289, 2014
|
|
3G9N
 
 | |
8D6U
 
 | | Rana catesbeiana saxiphilin:F-STX (soaked) | | Descriptor: | (2P)-4-({6-[({[(3aS,4R,7R,10aS)-2,6-diamino-10,10-dihydroxy-3a,4,9,10-tetrahydro-3H,8H-pyrrolo[1,2-c]purin-4-yl]methoxy}carbonyl)amino]hexyl}carbamoyl)-2-{[4aP,9(9a)P]-6-hydroxy-3-oxo-3H-xanthen-9-yl}benzoic acid, Saxiphilin | | Authors: | Chen, Z, Zakrzewska, S, Minor, D.L. | | Deposit date: | 2022-06-06 | | Release date: | 2022-11-02 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Definition of a saxitoxin (STX) binding code enables discovery and characterization of the anuran saxiphilin family.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
5IIS
 
 | | Design, synthesis and structure activity relationship of potent pan-PIM kinase inhibitors derived from the pyridyl-amide scaffold | | Descriptor: | 3-amino-N-(2'-amino-6'-methyl[4,4'-bipyridin]-3-yl)-6-(2-fluorophenyl)pyridine-2-carboxamide, DI(HYDROXYETHYL)ETHER, Serine/threonine-protein kinase pim-1 | | Authors: | Bellamacina, C, Bussiere, D, Burger, M. | | Deposit date: | 2016-03-01 | | Release date: | 2016-04-06 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Design, synthesis and structure activity relationship of potent pan-PIM kinase inhibitors derived from the pyridyl carboxamide scaffold.
Bioorg.Med.Chem.Lett., 26, 2016
|
|
6ZPJ
 
 | | Crystal structure of the unconventional kinetochore protein Leishmania mexicana KKT4 coiled coil domain | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, IMIDAZOLE, Leishmania mexicana KKT4 | | Authors: | Ludzia, P, Lowe, E.D, Marciano, G, Mohammed, S, Redfield, C, Akiyoshi, B. | | Deposit date: | 2020-07-08 | | Release date: | 2020-10-21 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural characterization of KKT4, an unconventional microtubule-binding kinetochore protein.
Structure, 29, 2021
|
|
8FLX
 
 | |
2MBO
 
 | |
9CJB
 
 | |
9CJF
 
 | |
