5A5D
 
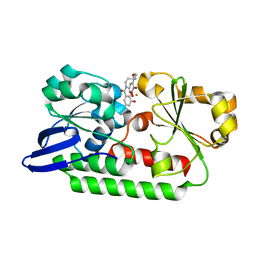 | | A complex of the synthetic siderophore analogue Fe(III)-5-LICAM with the CeuE periplasmic protein from Campylobacter jejuni | | Descriptor: | ENTEROCHELIN UPTAKE PERIPLASMIC BINDING PROTEIN, FE (III) ION, N,N'-pentane-1,5-diylbis(2,3-dihydroxybenzamide) | | Authors: | Blagova, E, Hughes, A, Moroz, O.V, Raines, D.J, Wilde, E.J, Turkenburg, J.P, Duhme-Klair, A.-K, Wilson, K.S. | | Deposit date: | 2015-06-17 | | Release date: | 2016-06-29 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Interactions of the periplasmic binding protein CeuE with Fe(III) n-LICAM(4-) siderophore analogues of varied linker length.
Sci Rep, 7, 2017
|
|
1AL2
 
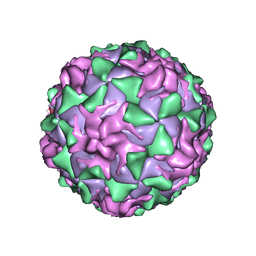 | | P1/MAHONEY POLIOVIRUS, SINGLE SITE MUTANT V1160I | | Descriptor: | MYRISTIC ACID, P1/MAHONEY POLIOVIRUS, SPHINGOSINE | | Authors: | Wien, M.W, Curry, S, Filman, D.J, Hogle, J.M. | | Deposit date: | 1997-06-09 | | Release date: | 1997-11-19 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural studies of poliovirus mutants that overcome receptor defects.
Nat.Struct.Biol., 4, 1997
|
|
1A72
 
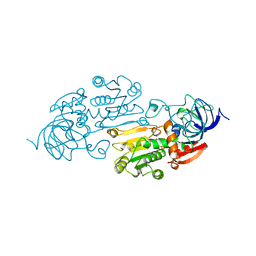 | | AN ACTIVE-SITE DOUBLE MUTANT (PHE93->TRP, VAL203->ALA) OF HORSE LIVER ALCOHOL DEHYDROGENASE IN COMPLEX WITH THE ISOSTERIC NAD ANALOG CPAD | | Descriptor: | 5-BETA-D-RIBOFURANOSYLPICOLINAMIDE ADENINE-DINUCLEOTIDE, HORSE LIVER ALCOHOL DEHYDROGENASE, ZINC ION | | Authors: | Colby, T.D, Bahnson, B.J, Chin, J.K, Klinman, J.P, Goldstein, B.M. | | Deposit date: | 1998-03-19 | | Release date: | 1998-06-17 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Active site modifications in a double mutant of liver alcohol dehydrogenase: structural studies of two enzyme-ligand complexes.
Biochemistry, 37, 1998
|
|
152D
 
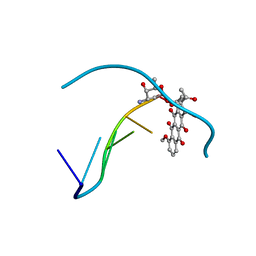 | | DIVERSITY OF WATER RING SIZE AT DNA INTERFACES: HYDRATION AND DYNAMICS OF DNA-ANTHRACYCLINE COMPLEXES | | Descriptor: | DAUNOMYCIN, DNA (5'-D(*CP*GP*AP*TP*CP*G)-3') | | Authors: | Lipscomb, L.A, Peek, M.E, Zhou, F.X, Bertrand, J.A, VanDerveer, D, Williams, L.D. | | Deposit date: | 1993-12-13 | | Release date: | 1994-05-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Water ring structure at DNA interfaces: hydration and dynamics of DNA-anthracycline complexes.
Biochemistry, 33, 1994
|
|
1A7G
 
 | |
1A5Y
 
 | |
4U2Z
 
 | | X-ray crystal structure of an Sco GlgEI-V279S/1,2,2-trifluromaltose complex | | Descriptor: | Alpha-1,4-glucan:maltose-1-phosphate maltosyltransferase 1, alpha-D-glucopyranose-(1-4)-2-deoxy-2,2-difluoro-alpha-D-arabino-hexopyranosyl fluoride | | Authors: | Ronning, D.R, Lindenberger, J.J. | | Deposit date: | 2014-07-18 | | Release date: | 2015-07-22 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Synthesis of 2-deoxy-2,2-difluoro-alpha-maltosyl fluoride and its X-ray structure in complex with Streptomyces coelicolor GlgEI-V279S.
Org.Biomol.Chem., 13, 2015
|
|
1AI2
 
 | | ISOCITRATE DEHYDROGENASE COMPLEXED WITH ISOCITRATE, NADP+, AND CALCIUM (FLASH-COOLED) | | Descriptor: | ISOCITRATE CALCIUM COMPLEX, ISOCITRATE DEHYDROGENASE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Stoddard, B.L, Mesecar, A, Koshland Junior, D.E. | | Deposit date: | 1997-04-30 | | Release date: | 1997-10-15 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Orbital steering in the catalytic power of enzymes: small structural changes with large catalytic consequences.
Science, 277, 1997
|
|
1AMP
 
 | | CRYSTAL STRUCTURE OF AEROMONAS PROTEOLYTICA AMINOPEPTIDASE: A PROTOTYPICAL MEMBER OF THE CO-CATALYTIC ZINC ENZYME FAMILY | | Descriptor: | AMINOPEPTIDASE, ZINC ION | | Authors: | Chevrier, B, Schalk, C, D'Orchymont, H, Rondeau, J.M, Moras, D, Tarnus, C. | | Deposit date: | 1994-04-22 | | Release date: | 1994-08-31 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of Aeromonas proteolytica aminopeptidase: a prototypical member of the co-catalytic zinc enzyme family.
Structure, 2, 1994
|
|
1AMJ
 
 | |
4UB3
 
 | | DNA polymerase beta closed product complex with a templating cytosine and 8-oxodGMP, 60 s | | Descriptor: | 5'-D(*CP*CP*GP*AP*CP*CP*GP*CP*GP*CP*AP*TP*CP*AP*GP*C)-3', 5'-D(*GP*CP*TP*GP*AP*TP*GP*CP*GP*CP*(8OG))-3', 5'-D(P*GP*TP*CP*GP*G)-3', ... | | Authors: | Freudenthal, B.D, Wilson, S.H, Beard, W.A. | | Deposit date: | 2014-08-11 | | Release date: | 2014-11-12 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Uncovering the polymerase-induced cytotoxicity of an oxidized nucleotide.
Nature, 517, 2015
|
|
4UBB
 
 | | DNA polymerase beta reactant complex with a templating cytosine and incoming 8-oxodGTP, 40 s | | Descriptor: | 5'-D(*CP*CP*GP*AP*CP*CP*GP*CP*GP*CP*AP*TP*CP*AP*GP*C)-3', 5'-D(*GP*CP*TP*GP*AP*TP*GP*CP*GP*CP*(8OG))-3', 5'-D(P*GP*TP*CP*GP*G)-3', ... | | Authors: | Freudenthal, B.D, Wilson, S.H, Beard, W.A. | | Deposit date: | 2014-08-12 | | Release date: | 2014-11-12 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Uncovering the polymerase-induced cytotoxicity of an oxidized nucleotide.
Nature, 517, 2015
|
|
1ADU
 
 | | EARLY E2A DNA-BINDING PROTEIN | | Descriptor: | ADENOVIRUS SINGLE-STRANDED DNA-BINDING PROTEIN, ZINC ION | | Authors: | Tucker, P.A, Kanellopoulos, P.N, Tsernoglou, D, Van Der Vliet, P.C. | | Deposit date: | 1995-05-11 | | Release date: | 1996-06-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Alternative arrangements of the protein chain are possible for the adenovirus single-stranded DNA binding protein.
J.Mol.Biol., 257, 1996
|
|
1A0D
 
 | | XYLOSE ISOMERASE FROM BACILLUS STEAROTHERMOPHILUS | | Descriptor: | MANGANESE (II) ION, XYLOSE ISOMERASE | | Authors: | Gallay, O, Chopra, R, Conti, E, Brick, P, Blow, D. | | Deposit date: | 1997-11-28 | | Release date: | 1998-06-03 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal Structures of Class II Xylose Isomerases from Two Thermophiles and a Hyperthermophile
To be Published
|
|
1AGX
 
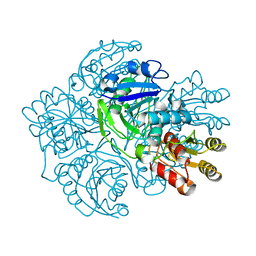 | | REFINED CRYSTAL STRUCTURE OF ACINETOBACTER GLUTAMINASIFICANS GLUTAMINASE-ASPARAGINASE | | Descriptor: | GLUTAMINASE-ASPARAGINASE | | Authors: | Lubkowski, J, Wlodawer, A, Housset, D, Weber, I.T, Ammon, H.L, Murphy, K.C, Swain, A.L. | | Deposit date: | 1994-07-13 | | Release date: | 1994-12-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Refined crystal structure of Acinetobacter glutaminasificans glutaminase-asparaginase.
Acta Crystallogr.,Sect.D, 50, 1994
|
|
4UAY
 
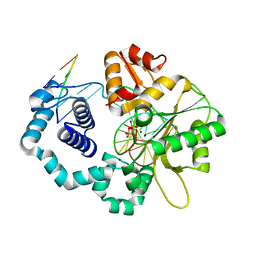 | | DNA polymerase beta product complex with a templating adenine and inserted 8-oxodGMP, 40 s | | Descriptor: | 5'-D(*CP*CP*GP*AP*CP*AP*GP*CP*GP*CP*AP*TP*CP*AP*GP*C)-3', 5'-D(*GP*CP*TP*GP*AP*TP*GP*CP*GP*CP*(8OG))-3', 5'-D(P*GP*TP*CP*GP*G)-3', ... | | Authors: | Freudenthal, B.D, Wilson, S.H, Beard, W.A. | | Deposit date: | 2014-08-11 | | Release date: | 2014-11-12 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.984 Å) | | Cite: | Uncovering the polymerase-induced cytotoxicity of an oxidized nucleotide.
Nature, 517, 2015
|
|
1AFL
 
 | |
1A99
 
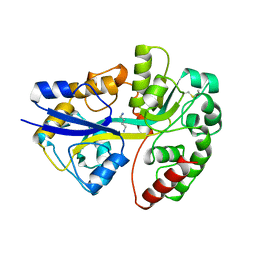 | | PUTRESCINE RECEPTOR (POTF) FROM E. COLI | | Descriptor: | 1,4-DIAMINOBUTANE, PUTRESCINE-BINDING PROTEIN | | Authors: | Vassylyev, D.G, Tomitori, H, Kashiwagi, K, Morikawa, K, Igarashi, K. | | Deposit date: | 1998-04-17 | | Release date: | 1998-10-21 | | Last modified: | 2020-04-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure and mutational analysis of the Escherichia coli putrescine receptor. Structural basis for substrate specificity.
J.Biol.Chem., 273, 1998
|
|
4UB4
 
 | | DNA polymerase beta substrate complex with a templating cytosine and incoming dGTP, 0 s | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, 5'-D(*CP*CP*GP*AP*CP*CP*GP*CP*GP*CP*AP*TP*CP*AP*GP*C)-3', 5'-D(*GP*CP*TP*GP*AP*TP*GP*CP*GP*C)-3', ... | | Authors: | Freudenthal, B.D, Wilson, S.H, Beard, W.A. | | Deposit date: | 2014-08-11 | | Release date: | 2014-11-12 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.954 Å) | | Cite: | Uncovering the polymerase-induced cytotoxicity of an oxidized nucleotide.
Nature, 517, 2015
|
|
1AJ2
 
 | | CRYSTAL STRUCTURE OF A BINARY COMPLEX OF E. COLI DIHYDROPTEROATE SYNTHASE | | Descriptor: | DIHYDROPTEROATE SYNTHASE, SULFATE ION, [7,8-DIHYDRO-PTERIN-6-YL METHANYL]-PHOSPHONOPHOSPHATE | | Authors: | Achari, A, Somers, D.O, Champness, J.N, Bryant, P.K, Rosemond, J, Stammers, D.K. | | Deposit date: | 1997-05-14 | | Release date: | 1998-05-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the anti-bacterial sulfonamide drug target dihydropteroate synthase.
Nat.Struct.Biol., 4, 1997
|
|
4UAW
 
 | | DNA polymerase beta substrate complex with a templating adenine and incoming 8-oxodGTP, 0 s | | Descriptor: | 5'-D(*CP*CP*GP*AP*CP*AP*GP*CP*GP*CP*AP*TP*CP*AP*GP*C)-3', 5'-D(*GP*CP*TP*GP*AP*TP*GP*CP*GP*C)-3', 5'-D(P*GP*TP*CP*GP*G)-3', ... | | Authors: | Freudenthal, B.D, Wilson, S.H, Beard, W.A. | | Deposit date: | 2014-08-11 | | Release date: | 2014-11-12 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Uncovering the polymerase-induced cytotoxicity of an oxidized nucleotide.
Nature, 517, 2015
|
|
4UB5
 
 | | DNA polymerase beta substrate complex with a templating cytosine, incoming 8-oxodGTP, and Mn2+, 5 s | | Descriptor: | 5'-D(*CP*CP*GP*AP*CP*CP*GP*CP*GP*CP*AP*TP*CP*AP*GP*C)-3', 5'-D(*GP*CP*TP*GP*AP*TP*GP*CP*GP*C)-3', 5'-D(P*GP*TP*CP*GP*G)-3', ... | | Authors: | Freudenthal, B.D, Wilson, S.H, Beard, W.A. | | Deposit date: | 2014-08-11 | | Release date: | 2014-11-12 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Uncovering the polymerase-induced cytotoxicity of an oxidized nucleotide.
Nature, 517, 2015
|
|
1A0N
 
 | | NMR STUDY OF THE SH3 DOMAIN FROM FYN PROTO-ONCOGENE TYROSINE KINASE COMPLEXED WITH THE SYNTHETIC PEPTIDE P2L CORRESPONDING TO RESIDUES 91-104 OF THE P85 SUBUNIT OF PI3-KINASE, FAMILY OF 25 STRUCTURES | | Descriptor: | FYN, PRO-PRO-ARG-PRO-LEU-PRO-VAL-ALA-PRO-GLY-SER-SER-LYS-THR | | Authors: | Renzoni, D.A, Pugh, D.J.R, Siligardi, G, Das, P, Morton, C.J, Rossi, C, Waterfield, M.D, Campbell, I.D, Ladbury, J.E. | | Deposit date: | 1997-12-05 | | Release date: | 1998-02-25 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural and thermodynamic characterization of the interaction of the SH3 domain from Fyn with the proline-rich binding site on the p85 subunit of PI3-kinase.
Biochemistry, 35, 1996
|
|
4UMM
 
 | | The Cryo-EM structure of the palindromic DNA-bound USP-EcR nuclear receptor reveals an asymmetric organization with allosteric domain positioning | | Descriptor: | 2,3,14,20,22-PENTAHYDROXYCHOLEST-7-EN-6-ONE, 5'-D(*CP*AP*AP*GP*GP*GP*TP*TP*CP*AP*AP*TP*GP*CP *AP*CP*TP*TP*GP*TP)-3', 5'-D(*DGP*AP*CP*AP*AP*GP*TP*GP*CP*AP*TP*TP*GP*DAP *AP*CP*CP*CP*TP*T)-3', ... | | Authors: | Maletta, M, Orlov, I, Moras, D, Billas, I.M.L, Klaholz, B.P. | | Deposit date: | 2014-05-19 | | Release date: | 2014-06-25 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (11.6 Å) | | Cite: | The Palindromic DNA-Bound Usp-Ecr Nuclear Receptor Adopts an Asymmetric Organization with Allosteric Domain Positioning.
Nat.Commun., 5, 2014
|
|
4UB1
 
 | | DNA polymerase beta product complex with a templating adenine and 8-oxodGMP, 90 s | | Descriptor: | 5'-D(*CP*CP*GP*AP*CP*AP*GP*CP*GP*CP*AP*TP*CP*AP*GP*C)-3', 5'-D(*GP*CP*TP*GP*AP*TP*GP*CP*GP*CP*(8OG))-3', 5'-D(P*GP*TP*CP*GP*G)-3', ... | | Authors: | Freudenthal, B.D, Wilson, S.H, Beard, W.A. | | Deposit date: | 2014-08-11 | | Release date: | 2014-11-12 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.341 Å) | | Cite: | Uncovering the polymerase-induced cytotoxicity of an oxidized nucleotide.
Nature, 517, 2015
|
|
