4Y63
 
 | | AAGlyB in complex with amino-acid analogues | | Descriptor: | 5'-deoxy-5'-{[(2S)-2-(triaza-1,2-dien-2-ium-1-yl)propanoyl]amino}uridine, Histo-blood group ABO system transferase, MANGANESE (II) ION, ... | | Authors: | Wang, S, Cuesta-Seijo, J.A, Striebeck, A, Lafont, D, Palcic, M.M, Vidal, S. | | Deposit date: | 2015-02-12 | | Release date: | 2015-09-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Design of Glycosyltransferase Inhibitors: Serine Analogues as Pyrophosphate Surrogates?
Chempluschem, 80, 2015
|
|
6YA9
 
 | | Crystal structure of rsGCaMP in the ON state (non-illuminated) | | Descriptor: | CALCIUM ION, rsCGaMP | | Authors: | Janowski, R, Fuenzalida-Werner, J.P, Mishra, K, Stiel, A.C, Niessing, D. | | Deposit date: | 2020-03-11 | | Release date: | 2021-10-06 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Genetically encoded photo-switchable molecular sensors for optoacoustic and super-resolution imaging.
Nat.Biotechnol., 2021
|
|
7AF5
 
 | | Bacterial 30S ribosomal subunit assembly complex state I (head domain) | | Descriptor: | 16SrRNA (head domain of the 30S ribosome), 30S ribosomal protein S10, 30S ribosomal protein S13, ... | | Authors: | Schedlbauer, A, Iturrioz, I, Ochoa-Lizarralde, B, Diercks, T, Lopez-Alonso, J, Kaminishi, T, Capuni, R, Astigarraga, E, Gil-Carton, D, Fucini, P, Connell, S. | | Deposit date: | 2020-09-19 | | Release date: | 2021-07-07 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.96 Å) | | Cite: | A conserved rRNA switch is central to decoding site maturation on the small ribosomal subunit.
Sci Adv, 7, 2021
|
|
4YI8
 
 | |
6N21
 
 | | Structure of wild-type CAO1 | | Descriptor: | CHLORIDE ION, Carotenoid oxygenase, FE (II) ION | | Authors: | Khadka, N, Kiser, P.D. | | Deposit date: | 2018-11-12 | | Release date: | 2019-06-05 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Evidence for distinct rate-limiting steps in the cleavage of alkenes by carotenoid cleavage dioxygenases.
J.Biol.Chem., 294, 2019
|
|
7KWW
 
 | | X-ray Crystal Structure of PlyCB Mutant K59H | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, PlyCB | | Authors: | Williams, D.E, Broendum, S.S, Hayes, B.K, Drinkwater, N, McGowan, S. | | Deposit date: | 2020-12-02 | | Release date: | 2021-04-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | High avidity drives the interaction between the streptococcal C1 phage endolysin, PlyC, with the cell surface carbohydrates of Group A Streptococcus.
Mol.Microbiol., 116, 2021
|
|
6N1Y
 
 | | Structure of L509V CAO1 - growth condition 1 | | Descriptor: | CHLORIDE ION, Carotenoid oxygenase, FE (II) ION | | Authors: | Khadka, N, Kiser, P.D. | | Deposit date: | 2018-11-12 | | Release date: | 2019-06-05 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Evidence for distinct rate-limiting steps in the cleavage of alkenes by carotenoid cleavage dioxygenases.
J.Biol.Chem., 294, 2019
|
|
7KWY
 
 | | X-ray Crystal Structure of PlyCB Mutant R66K | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, PlyCB | | Authors: | Williams, D.E, Broendum, S.S, Hayes, B.K, Drinkwater, N, McGowan, S. | | Deposit date: | 2020-12-02 | | Release date: | 2021-04-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | High avidity drives the interaction between the streptococcal C1 phage endolysin, PlyC, with the cell surface carbohydrates of Group A Streptococcus.
Mol.Microbiol., 116, 2021
|
|
6PVA
 
 | | The structure of NTMT1 in complex with compound 11 | | Descriptor: | AMINO GROUP-()-LYSINE-()-LYSINE-()-PROLINE-()-AMINO-ACETALDEHYDE-()-5'-{[(3S)-3-amino-3-carboxypropyl](3-aminopropyl)amino}-5'-deoxyadenosine, N-terminal Xaa-Pro-Lys N-methyltransferase 1 | | Authors: | Noinaj, N, Chen, D, Huang, R. | | Deposit date: | 2019-07-20 | | Release date: | 2020-08-19 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | The structure of NTMT1 in complex with compound 11
To Be published
|
|
2VJB
 
 | | Torpedo Californica Acetylcholinesterase In Complex With A Non Hydrolysable Substrate Analogue, 4-Oxo-N,N,N- Trimethylpentanaminium - Orthorhombic space group - Dataset D at 100K | | Descriptor: | (4R)-4-HYDROXY-N,N,N-TRIMETHYLPENTAN-1-AMINIUM, 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETYLCHOLINESTERASE, ... | | Authors: | Colletier, J.P, Bourgeois, D, Fournier, D, Silman, I, Sussman, J.L, Weik, M. | | Deposit date: | 2007-12-09 | | Release date: | 2008-07-22 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Shoot-and-Trap: Use of Specific X-Ray Damage to Study Structural Protein Dynamics by Temperature-Controlled Cryo-Crystallography.
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
6PVJ
 
 | | Crystal structure of PhqK in complex with malbrancheamide C | | Descriptor: | (5aS,12aS,13aS)-9-bromo-12,12-dimethyl-2,3,11,12,12a,13-hexahydro-1H,5H,6H-5a,13a-(epiminomethano)indolizino[7,6-b]carbazol-14-one, FAD monooxygenase, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Fraley, A.E, Smith, J.L, Sherman, D.H. | | Deposit date: | 2019-07-20 | | Release date: | 2020-01-22 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Molecular Basis for Spirocycle Formation in the Paraherquamide Biosynthetic Pathway.
J.Am.Chem.Soc., 142, 2020
|
|
7KWT
 
 | | X-ray Crystal Structure of PlyCB Mutant Y28H | | Descriptor: | PlyCB | | Authors: | Williams, D.E, Broendum, S.S, Hayes, B.K, Drinkwater, N, McGowan, S. | | Deposit date: | 2020-12-02 | | Release date: | 2021-04-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | High avidity drives the interaction between the streptococcal C1 phage endolysin, PlyC, with the cell surface carbohydrates of Group A Streptococcus.
Mol.Microbiol., 116, 2021
|
|
6Q1J
 
 | |
5GS5
 
 | | Crystal structure of apo rat STING | | Descriptor: | SULFATE ION, Stimulator of interferon genes protein | | Authors: | Zhang, H, Han, M.J, Tao, J.L, Ye, Z.Y, Du, X.X, Deng, M.J, Zhang, X.Y, Li, L.F, Jiang, Z.F, Su, X.D. | | Deposit date: | 2016-08-13 | | Release date: | 2017-10-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Crystal structure of apo ratSTING
To Be Published
|
|
4RLL
 
 | | Crystal structure of human CK2alpha in complex with the ATP-competitive inhibitor 4-[(E)-(fluoren-9-ylidenehydrazinylidene)-methyl] benzoate | | Descriptor: | 4-[(E)-(9H-fluoren-9-ylidenehydrazinylidene)methyl]benzoic acid, Casein kinase II subunit alpha, GLYCEROL | | Authors: | Guerra, B, Rasmussen, T.D.L, Schnitzler, A, Jensen, H.H, Boldyreff, B.S, Miyata, Y, Marcussen, N, Niefind, K, Issinger, O.G. | | Deposit date: | 2014-10-17 | | Release date: | 2014-12-17 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Protein kinase CK2 inhibition is associated with the destabilization of HIF-1 alpha in human cancer cells.
Cancer Lett, 356, 2015
|
|
4YSY
 
 | | Crystal structure of Mitochondrial rhodoquinol-fumarate reductase from Ascaris suum with N-[(2,4-dichlorophenyl)methyl]-2-(trifluoromethyl)benzamide | | Descriptor: | Cytochrome b-large subunit, FE2/S2 (INORGANIC) CLUSTER, FE3-S4 CLUSTER, ... | | Authors: | Harada, S, Shiba, T, Sato, D, Yamamoto, A, Nagahama, M, Yone, A, Inaoka, D.K, Sakamoto, K, Inoue, M, Honma, T, Kita, K. | | Deposit date: | 2015-03-17 | | Release date: | 2015-08-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural Insights into the Molecular Design of Flutolanil Derivatives Targeted for Fumarate Respiration of Parasite Mitochondria
Int J Mol Sci, 16, 2015
|
|
6FCM
 
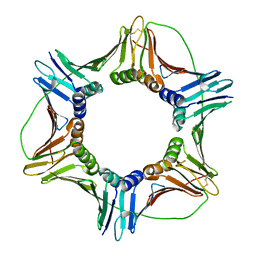 | | Crystal structure of human PCNA | | Descriptor: | Proliferating cell nuclear antigen | | Authors: | Housset, D, Frachet, P. | | Deposit date: | 2017-12-20 | | Release date: | 2019-01-30 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Cytosolic PCNA interacts with p47phox and controls NADPH oxidase NOX2 activation in neutrophils.
J.Exp.Med., 216, 2019
|
|
6YZ0
 
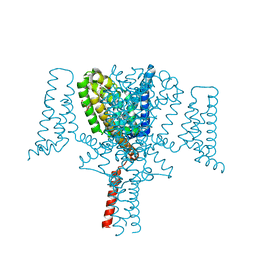 | | Full length Open-form Sodium Channel NavMs F208L in complex with Cannabidiol (CBD) | | Descriptor: | 2-[(1R,2R,5S)-5-methyl-2-(prop-1-en-2-yl)cyclohexyl]-5-pentylbenzene-1,3-diol, DODECAETHYLENE GLYCOL, HEGA-10, ... | | Authors: | Sula, A, Sait, L.G, Hollingworth, D, Wallace, B.A. | | Deposit date: | 2020-05-06 | | Release date: | 2020-11-11 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Cannabidiol interactions with voltage-gated sodium channels.
Elife, 9, 2020
|
|
8KF4
 
 | |
4G59
 
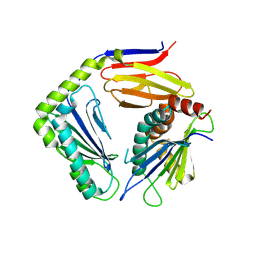 | |
5NHI
 
 | |
4G6B
 
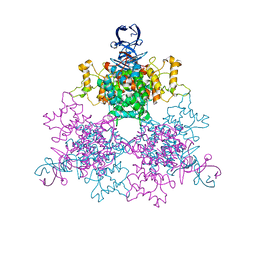 | | Three dimensional structure analysis of the type II citrate synthase from e.coli | | Descriptor: | Citrate synthase, SULFATE ION | | Authors: | Nguyen, N.T, Maurus, R, Stokell, D.J, Ayed, A, Duckworth, H.W, Brayer, G.D. | | Deposit date: | 2012-07-18 | | Release date: | 2013-11-20 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Comparative Analysis of Folding and Substrate Binding Sites between Regulated Hexameric Type II Citrate Synthases and Unregulated Dimeric Type I Enzymes.
Biochemistry, 40, 2001
|
|
5U8W
 
 | | Dihydrolipoamide dehydrogenase (LpdG) from Pseudomonas aeruginosa bound to NADH | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, DIMETHYL SULFOXIDE, Dihydrolipoyl dehydrogenase, ... | | Authors: | Glasser, N.R, Wang, B.X, Hoy, J.A, Newman, D.K. | | Deposit date: | 2016-12-15 | | Release date: | 2017-02-15 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | The Pyruvate and alpha-Ketoglutarate Dehydrogenase Complexes of Pseudomonas aeruginosa Catalyze Pyocyanin and Phenazine-1-carboxylic Acid Reduction via the Subunit Dihydrolipoamide Dehydrogenase.
J. Biol. Chem., 292, 2017
|
|
6BAW
 
 | | Structure of GRP94 with a selective resorcinylic inhibitor. | | Descriptor: | 1,2-ETHANEDIOL, Endoplasmin, PHOSPHATE ION, ... | | Authors: | Que, N.L.S, Gewirth, D.T. | | Deposit date: | 2017-10-16 | | Release date: | 2018-04-18 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.703 Å) | | Cite: | Structure Based Design of a Grp94-Selective Inhibitor: Exploiting a Key Residue in Grp94 To Optimize Paralog-Selective Binding.
J. Med. Chem., 61, 2018
|
|
4Y62
 
 | | AAGlyB in complex with amino-acid analogues | | Descriptor: | 5'-deoxy-5'-{[(2R)-3-hydroxy-2-(4-phenyl-1H-1,2,3-triazol-1-yl)propanoyl]amino}uridine, Histo-blood group ABO system transferase, octyl 2-O-(6-deoxy-alpha-L-galactopyranosyl)-beta-D-galactopyranoside | | Authors: | Wang, S, Cuesta-Seijo, J.A, Striebeck, A, Lafont, D, Palcic, M.M, Vidal, S. | | Deposit date: | 2015-02-12 | | Release date: | 2015-09-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Design of Glycosyltransferase Inhibitors: Serine Analogues as Pyrophosphate Surrogates?
Chempluschem, 80, 2015
|
|
