5X7F
 
 | |
8PPO
 
 | |
6QSG
 
 | | Crystal structure of the hybrid bioinorganic complex of Pizza6S and Keggin (STA) | | Descriptor: | Keggin (STA), Pizza6S | | Authors: | Noguchi, H, Vandebroek, L, Kamata, K, Tame, J.R.H, Van Meervelt, L, Parac-Vogt, T.N, Voet, A.R.D. | | Deposit date: | 2019-02-20 | | Release date: | 2020-03-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Hybrid assemblies of a symmetric designer protein and polyoxometalates with matching symmetry.
Chem.Commun.(Camb.), 56, 2020
|
|
8T10
 
 | | TRPV1 in nanodisc bound with two LPA molecules in opposite monomers | | Descriptor: | (2R)-2-hydroxy-3-(phosphonooxy)propyl tetradecanoate, (2R)-3-{[(R)-hydroxy{[(1S,2R,3R,4S,5S,6R)-2,3,4,5,6-pentahydroxycyclohexyl]oxy}phosphoryl]oxy}propane-1,2-diyl dioctadecanoate, SODIUM ION, ... | | Authors: | Arnold, W.R, Julius, D, Cheng, Y. | | Deposit date: | 2023-06-01 | | Release date: | 2024-05-08 | | Last modified: | 2024-10-02 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural basis of TRPV1 modulation by endogenous bioactive lipids.
Nat.Struct.Mol.Biol., 31, 2024
|
|
2IJR
 
 | | Crystal structure of a protein api92 from Yersinia pseudotuberculosis, Pfam DUF1281 | | Descriptor: | Hypothetical protein api92 | | Authors: | Jin, X, Min, T, Bonanno, J.B, Sauder, J.M, Wasserman, S, Smith, D, Burley, S.K, Shapiro, L, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-09-30 | | Release date: | 2006-10-31 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of a hypothetical protein from Yersinia
pseudotuberculosis
To be Published
|
|
1D32
 
 | | DRUG-INDUCED DNA REPAIR: X-RAY STRUCTURE OF A DNA-DITERCALINIUM COMPLEX | | Descriptor: | DITERCALINIUM, DNA (5'-D(*CP*GP*CP*G)-3') | | Authors: | Gao, Q, Williams, L.D, Egli, M, Rabinovich, D, Chen, S.-L, Quigley, G.J, Rich, A. | | Deposit date: | 1991-01-23 | | Release date: | 1992-04-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Drug-induced DNA repair: X-ray structure of a DNA-ditercalinium complex.
Proc.Natl.Acad.Sci.USA, 88, 1991
|
|
7LFB
 
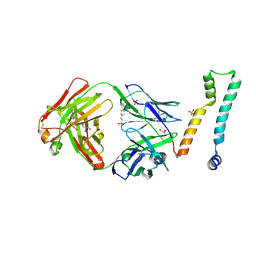 | | Fab 7D6 bound to ApoL1 NTD | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Apolipoprotein L1, Fab 7D6 heavy chain, ... | | Authors: | Ultsch, M, Kirchhofer, D. | | Deposit date: | 2021-01-16 | | Release date: | 2021-08-04 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.913 Å) | | Cite: | Structures of the ApoL1 and ApoL2 N-terminal domains reveal a non-classical four-helix bundle motif.
Commun Biol, 4, 2021
|
|
6QUK
 
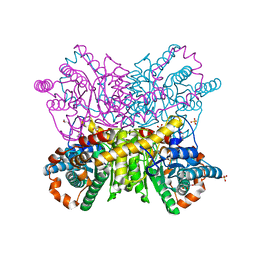 | | Protein crystallization by ionic liquid hydrogel support: glucose isomerase grown by using ionic liquid hydrogel | | Descriptor: | GLYCEROL, MANGANESE (II) ION, SULFATE ION, ... | | Authors: | Belviso, B.D, Caliandro, R, Caliandro, R. | | Deposit date: | 2019-02-27 | | Release date: | 2019-03-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Protein Crystallization in Ionic-Liquid Hydrogel Composite Membranes
Crystals, 9, 2019
|
|
6QZB
 
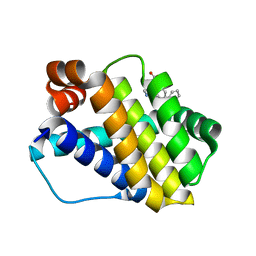 | | Structure of Mcl-1 in complex with compound 8d | | Descriptor: | (2~{R})-2-[[6-ethyl-5-(2-methylphenyl)thieno[2,3-d]pyrimidin-4-yl]amino]-3-phenyl-propanoic acid, Induced myeloid leukemia cell differentiation protein Mcl-1 | | Authors: | Dokurno, P, Szlavik, Z, Ondi, L, Csekei, M, Paczal, A, Szabo, Z.B, Radics, G, Murray, J, Davidson, J, Chen, I, Davis, B, Hubbard, R.E, Pedder, C, Surgenor, A.E, Smith, J, Robertson, A, LeToumelin-Braizat, G, Cauquil, N, Zarka, M, Demarles, D, Perron-Sierra, F, Geneste, O, Kotschy, A. | | Deposit date: | 2019-03-11 | | Release date: | 2019-08-07 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-Guided Discovery of a Selective Mcl-1 Inhibitor with Cellular Activity.
J.Med.Chem., 62, 2019
|
|
1D4M
 
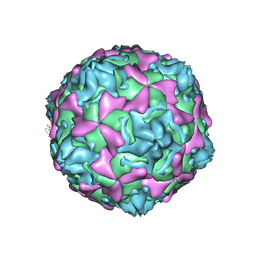 | | THE CRYSTAL STRUCTURE OF COXSACKIEVIRUS A9 TO 2.9 A RESOLUTION | | Descriptor: | 5-(7-(4-(4,5-DIHYDRO-2-OXAZOLYL)PHENOXY)HEPTYL)-3-METHYL ISOXAZOLE, MYRISTIC ACID, PROTEIN (COXSACKIEVIRUS A9) | | Authors: | Hendry, E, Hatanaka, H, Fry, E, Smyth, M, Tate, J, Stanway, G, Santti, J, Maaronen, M, Hyypia, T, Stuart, D. | | Deposit date: | 1999-10-04 | | Release date: | 1999-12-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The crystal structure of coxsackievirus A9: new insights into the uncoating mechanisms of enteroviruses.
Structure Fold.Des., 7, 1999
|
|
5KX1
 
 | |
1D6O
 
 | | NATIVE FKBP | | Descriptor: | AMMONIUM ION, PROTEIN (FK506-BINDING PROTEIN), SULFATE ION | | Authors: | Burkhard, P, Taylor, P, Walkinshaw, M.D. | | Deposit date: | 1999-10-15 | | Release date: | 1999-10-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | X-ray structures of small ligand-FKBP complexes provide an estimate for hydrophobic interaction energies.
J.Mol.Biol., 295, 2000
|
|
7LF7
 
 | | Fab 6D12 bound to ApoL1 NTD | | Descriptor: | Apolipoprotein L1, ETHANOL, Fab 6D12 heavy chain, ... | | Authors: | Ultsch, M, Kirchhofer, D. | | Deposit date: | 2021-01-15 | | Release date: | 2021-08-04 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.026 Å) | | Cite: | Structures of the ApoL1 and ApoL2 N-terminal domains reveal a non-classical four-helix bundle motif.
Commun Biol, 4, 2021
|
|
7L5X
 
 | | p97-R155H mutant dodecamer II | | Descriptor: | Transitional endoplasmic reticulum ATPase | | Authors: | Nandi, P, Li, S, Coulmbres, R.C.A, Wang, F, Williams, D.R, Malyutin, A.G, Poh, Y.-P, Chou, T.-F, Chiu, P.-L. | | Deposit date: | 2020-12-23 | | Release date: | 2021-08-04 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (6.1 Å) | | Cite: | Structural and Functional Analysis of Disease-Linked p97 ATPase Mutant Complexes.
Int J Mol Sci, 22, 2021
|
|
5ZYJ
 
 | | Crystal structure of CERT START domain in complex with compound E16A | | Descriptor: | 2-[4-[4-pentyl-3-[(1~{S},2~{R})-2-pyridin-2-ylcyclopropyl]phenyl]phenyl]sulfonylethanol, GLYCEROL, LIPID-TRANSFER PROTEIN CERT | | Authors: | Suzuki, M, Nakao, N, Ueno, M, Sakai, S, Egawa, D, Hanzawa, H, Kawasaki, S, Kumagai, K, Kobayashi, S, Hanada, K. | | Deposit date: | 2018-05-25 | | Release date: | 2019-02-27 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Natural ligand-nonmimetic inhibitors of the lipid-transfer protein CERT
Commun Chem, 2019
|
|
6X2Z
 
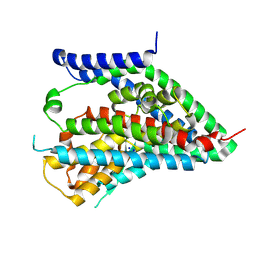 | | hEAAT3-OFS-Asp | | Descriptor: | ASPARTIC ACID, Excitatory amino acid transporter 3, SODIUM ION | | Authors: | Qiu, B, Matthies, D, Boudker, O. | | Deposit date: | 2020-05-21 | | Release date: | 2021-03-17 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.03 Å) | | Cite: | Cryo-EM structures of excitatory amino acid transporter 3 visualize coupled substrate, sodium, and proton binding and transport.
Sci Adv, 7, 2021
|
|
7LFD
 
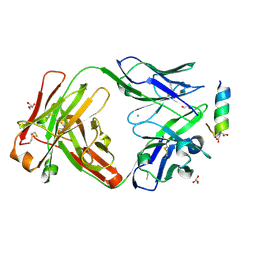 | | Fab 7D6 bound to ApoL1 BH3 like peptide | | Descriptor: | AMMONIUM ION, Apolipoprotein L1 BH3 like peptide, CITRATE ANION, ... | | Authors: | Ultsch, M, Kirchhofer, D. | | Deposit date: | 2021-01-16 | | Release date: | 2021-08-04 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.157 Å) | | Cite: | Structures of the ApoL1 and ApoL2 N-terminal domains reveal a non-classical four-helix bundle motif.
Commun Biol, 4, 2021
|
|
3EOE
 
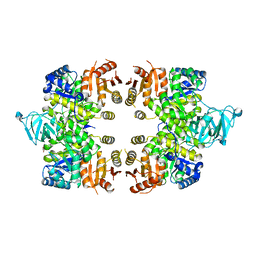 | | Crystal Structure of Pyruvate Kinase from toxoplasma gondii, 55.m00007 | | Descriptor: | GLYCEROL, Pyruvate kinase | | Authors: | Wernimont, A.K, Lew, J, Kozieradzki, I, Wasney, G, Hassani, A, Vedadi, M, Cossar, D, Schapiro, M, Bochkarev, A, Arrowsmith, C.H, Bountra, C, Weigelt, J, Edwards, A.M, Hui, R, Pizarro, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-09-26 | | Release date: | 2008-10-14 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | The crystal structure of Toxoplasma gondii pyruvate kinase 1.
Plos One, 5, 2010
|
|
6X3E
 
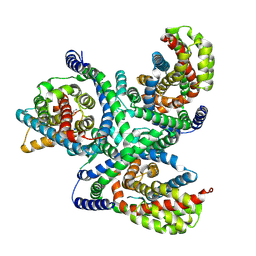 | | hEAAT3-Asymmetric-1o2i | | Descriptor: | ASPARTIC ACID, Excitatory amino acid transporter 3, SODIUM ION | | Authors: | Qiu, B, Matthies, D, Boudker, O. | | Deposit date: | 2020-05-21 | | Release date: | 2021-03-17 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.42 Å) | | Cite: | Cryo-EM structures of excitatory amino acid transporter 3 visualize coupled substrate, sodium, and proton binding and transport.
Sci Adv, 7, 2021
|
|
157D
 
 | | CRYSTAL AND MOLECULAR STRUCTURE OF R(CGCGAAUUAGCG): AN RNA DUPLEX CONTAINING TWO G(ANTI).A(ANTI) BASE-PAIRS | | Descriptor: | RNA (5'-R(*CP*GP*CP*GP*AP*AP*UP*UP*AP*GP*CP*G)-3') | | Authors: | Leonard, G.A, McAuley-Hecht, K.E, Ebel, S, Lough, D.M, Brown, T, Hunter, W.N. | | Deposit date: | 1994-02-01 | | Release date: | 1994-05-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal and molecular structure of r(CGCGAAUUAGCG): an RNA duplex containing two G(anti).A(anti) base pairs.
Structure, 2, 1994
|
|
6WQU
 
 | | CSL (RBPJ) bound to Notch3 RAM and DNA | | Descriptor: | DNA (5'-D(*AP*AP*TP*CP*TP*TP*TP*CP*CP*CP*AP*CP*GP*GP*T)-3'), DNA (5'-D(*TP*TP*AP*CP*CP*GP*TP*GP*GP*GP*AP*AP*AP*GP*A)-3'), Neurogenic locus notch homolog protein 3, ... | | Authors: | Kovall, R.A, Gagliani, E, Hall, D. | | Deposit date: | 2020-04-29 | | Release date: | 2021-03-31 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | PIM-induced phosphorylation of Notch3 promotes breast cancer tumorigenicity in a CSL-independent fashion.
J.Biol.Chem., 296, 2021
|
|
5EPN
 
 | | Crystal structure of HCV NS3/4A protease in complex with 5172-mcP1P3 (MK-5172 P1-P3 macrocyclic analogue) | | Descriptor: | 2-Methyl-2-propanyl {(2R,6S,12Z,13aS,14aR,16aS)-14a-[(cyclopropylsulfonyl)carbamoyl]-2-[(3-ethyl-7-methoxy-2-quinoxalinyl)oxy]-5,16-dioxo-1,2,3,5,6,7,8,9,10,11,13a,14,14a,15,16,16a-hexadecahydrocyclop ropa[e]pyrrolo[1,2-a][1,4]diazacyclopentadecin-6-yl}carbamate, NS3 protease, SULFATE ION, ... | | Authors: | Soumana, D.I, Yilmaz, N.K, Ali, A, Prachanronarong, K.L, Aydin, C, Schiffer, C.A. | | Deposit date: | 2015-11-11 | | Release date: | 2016-01-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and Thermodynamic Effects of Macrocyclization in HCV NS3/4A Inhibitor MK-5172.
Acs Chem.Biol., 11, 2016
|
|
5XFJ
 
 | | Crystal structure of LY2874455 in complex of FGFR4 gatekeeper mutation (V550M) | | Descriptor: | 2-[4-[E-2-[5-[(1R)-1-[3,5-bis(chloranyl)pyridin-4-yl]ethoxy]-1H-indazol-3-yl]ethenyl]pyrazol-1-yl]ethanol, Fibroblast growth factor receptor 4 | | Authors: | Wu, D, Chen, Y. | | Deposit date: | 2017-04-10 | | Release date: | 2018-08-08 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | LY2874455 potently inhibits FGFR gatekeeper mutants and overcomes mutation-based resistance.
Chem. Commun. (Camb.), 54, 2018
|
|
6X3F
 
 | | hEAAT3-IFS-Apo | | Descriptor: | CHOLINE ION, Excitatory amino acid transporter 3 | | Authors: | Qiu, B, Matthies, D, Boudker, O. | | Deposit date: | 2020-05-21 | | Release date: | 2021-03-17 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.03 Å) | | Cite: | Cryo-EM structures of excitatory amino acid transporter 3 visualize coupled substrate, sodium, and proton binding and transport.
Sci Adv, 7, 2021
|
|
7L4U
 
 | | Crystal structure of human monoacylglycerol lipase in complex with compound 1h | | Descriptor: | (5S)-5-(3-{4-[(2-chloro-4-fluorophenoxy)methyl]piperidin-1-yl}-3-oxopropyl)pyrrolidin-2-one, CHLORIDE ION, Monoglyceride lipase | | Authors: | Qin, L, Lane, W, Skene, R.J, Dougan, D. | | Deposit date: | 2020-12-21 | | Release date: | 2021-08-11 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Design and Synthesis of Novel Spiro Derivatives as Potent and Reversible Monoacylglycerol Lipase (MAGL) Inhibitors: Bioisosteric Transformation from 3-Oxo-3,4-dihydro-2 H -benzo[ b ][1,4]oxazin-6-yl Moiety.
J.Med.Chem., 64, 2021
|
|
