5JRL
 
 | |
7NTD
 
 | | The structure of the SBP TarP_Csal in complex with ferulate | | Descriptor: | 3-(4-HYDROXY-3-METHOXYPHENYL)-2-PROPENOIC ACID, MAGNESIUM ION, SULFATE ION, ... | | Authors: | Bisson, C, Salmon, R.C, West, L, Rafferty, J.B, Hitchcock, A, Thomas, G.H, Kelly, D.J. | | Deposit date: | 2021-03-09 | | Release date: | 2021-10-06 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The structural basis for high-affinity uptake of lignin-derived aromatic compounds by proteobacterial TRAP transporters.
Febs J., 289, 2022
|
|
7YWP
 
 | | Closed conformation of Oligopeptidase B from Serratia proteomaculans with covalently bound TCK | | Descriptor: | N-[(1S)-5-amino-1-(chloroacetyl)pentyl]-4-methylbenzenesulfonamide, Oligopeptidase B | | Authors: | Petrenko, D.E, Boyko, K.M, Nikolaeva, A.Y, Vlaskina, A.V, Mikhailova, A.G, Timofeev, V.I, Rakitina, T.V. | | Deposit date: | 2022-02-14 | | Release date: | 2023-02-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of Inhibitor-Bound Bacterial Oligopeptidase B in the Closed State: Similarity and Difference between Protozoan and Bacterial Enzymes.
Int J Mol Sci, 24, 2023
|
|
6SCM
 
 | | SOS1 in Complex with Inhibitor BI-3406 | | Descriptor: | 1,2-ETHANEDIOL, IMIDAZOLE, Son of sevenless homolog 1, ... | | Authors: | Kessler, D, Fischer, G, Ramharter, J. | | Deposit date: | 2019-07-24 | | Release date: | 2020-08-26 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.866 Å) | | Cite: | BI-3406, a Potent and Selective SOS1-KRAS Interaction Inhibitor, Is Effective in KRAS-Driven Cancers through Combined MEK Inhibition.
Cancer Discov, 11, 2021
|
|
6FCN
 
 | |
7NTE
 
 | | The structure of an open conformation of the SBP TarP_Csal | | Descriptor: | MAGNESIUM ION, TRAP dicarboxylate transporter-DctP subunit | | Authors: | Bisson, C, Salmon, R.C, West, L, Rafferty, J.B, Hitchcock, A, Thomas, G.H, Kelly, D.J. | | Deposit date: | 2021-03-09 | | Release date: | 2021-10-06 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The structural basis for high-affinity uptake of lignin-derived aromatic compounds by proteobacterial TRAP transporters.
Febs J., 289, 2022
|
|
5MI9
 
 | | Structure of the phosphomimetic mutant of the elongation factor EF-Tu T62E | | Descriptor: | Elongation factor Tu 1, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION | | Authors: | Talavera, A, Hendrix, J, Versees, W, De Gieter, S, Castro-Roa, D, Jurenas, D, Van Nerom, K, Vandenberk, N, Barth, A, De Greve, H, Hofkens, J, Zenkin, N, Loris, R, Garcia-Pino, A. | | Deposit date: | 2016-11-27 | | Release date: | 2017-12-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Phosphorylation decelerates conformational dynamics in bacterial translation elongation factors.
Sci Adv, 4, 2018
|
|
6C97
 
 | | Crystal structure of FcRn at pH3 | | Descriptor: | Beta-2-microglobulin, GLYCEROL, IgG receptor FcRn large subunit p51 | | Authors: | Fox III, D, Fairman, J.W. | | Deposit date: | 2018-01-25 | | Release date: | 2018-05-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Insight into small molecule binding to the neonatal Fc receptor by X-ray crystallography and 100 kHz magic-angle-spinning NMR.
PLoS Biol., 16, 2018
|
|
6UW3
 
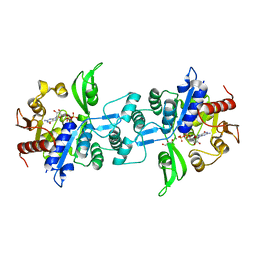 | | The crystal structure of FbiA from Mycobacterium Smegmatis, GDP Bound form | | Descriptor: | CALCIUM ION, GLYCEROL, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Grinter, R, Gillett, D, Cordero, P.R.F, Greening, C. | | Deposit date: | 2019-11-04 | | Release date: | 2020-05-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Cellular and Structural Basis of Synthesis of the Unique Intermediate Dehydro-F420-0 in Mycobacteria.
mSystems, 5, 2020
|
|
8H8Y
 
 | | Crystal structure of AbHheG from Acidimicrobiia bacterium | | Descriptor: | GLYCEROL, alpha/beta hydrolase | | Authors: | Zhou, C.H, Chen, X, Han, X, Liu, W.D, Wu, Q.Q, Zhu, D.M, Ma, Y.H. | | Deposit date: | 2022-10-24 | | Release date: | 2023-08-02 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Flipping the Substrate Creates a Highly Selective Halohydrin Dehalogenase for the Synthesis of Chiral 4-Aryl-2-oxazolidinones from Readily Available Epoxides
Acs Catalysis, 13, 2023
|
|
8H3C
 
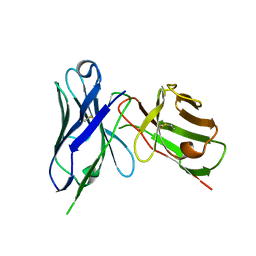 | | Crystal structure of M2e Influenza peptide in complex with antibody scFv | | Descriptor: | Matrix protein 2, Single Chain Variable Fragment | | Authors: | Kumar, U, Madni, Z.K, Gaur, V, Salunke, D.M. | | Deposit date: | 2022-10-08 | | Release date: | 2023-08-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.43 Å) | | Cite: | A structure and knowledge-based combinatorial approach to engineering universal scFv antibodies against influenza M2 protein.
J.Biomed.Sci., 30, 2023
|
|
7Q97
 
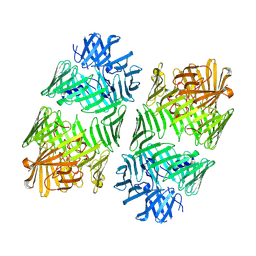 | | Structure of the bacterial type VI secretion system effector RhsA. | | Descriptor: | Rhs family protein | | Authors: | Guenther, P, Quentin, D, Ahmad, S, Sachar, K, Gatsogiannis, C, Whitney, J.C, Raunser, S. | | Deposit date: | 2021-11-12 | | Release date: | 2021-12-22 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structure of a bacterial Rhs effector exported by the type VI secretion system.
Plos Pathog., 18, 2022
|
|
4XXS
 
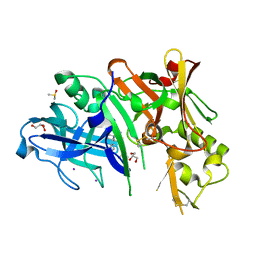 | | Crystal structure of BACE1 with a pyrazole-substituted tetrahydropyran thioamidine | | Descriptor: | (4aR,6R,8aS)-8a-(2,4-difluorophenyl)-6-(1-methyl-1H-pyrazol-4-yl)-4,4a,5,6,8,8a-hexahydropyrano[3,4-d][1,3]thiazin-2-amine, Beta-secretase 1, DIMETHYL SULFOXIDE, ... | | Authors: | Parris, K.D, Pandit, J. | | Deposit date: | 2015-01-30 | | Release date: | 2015-04-01 | | Last modified: | 2015-04-22 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Utilizing Structures of CYP2D6 and BACE1 Complexes To Reduce Risk of Drug-Drug Interactions with a Novel Series of Centrally Efficacious BACE1 Inhibitors.
J.Med.Chem., 58, 2015
|
|
8CEE
 
 | | Rnase R bound to a 30S degradation intermediate (State I - head-turning) | | Descriptor: | 16S rRNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Paternoga, H, Dimitrova-Paternoga, L, Wilson, D.N. | | Deposit date: | 2023-02-01 | | Release date: | 2023-12-20 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural basis of ribosomal 30S subunit degradation by RNase R.
Nature, 626, 2024
|
|
5MMR
 
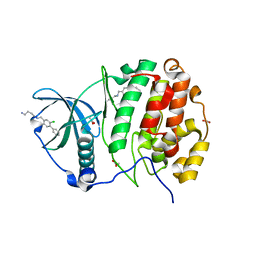 | | Crystal Structure of CK2alpha with N-((2-chloro-[1,1'-biphenyl]-4-yl)methyl)butane-1,4-diamine bound | | Descriptor: | ACETATE ION, Casein kinase II subunit alpha, PHOSPHATE ION, ... | | Authors: | Brear, P, De Fusco, C, Georgiou, K, Iegre, J, Sore, H, Hyvonen, M, Spring, D. | | Deposit date: | 2016-12-12 | | Release date: | 2017-05-24 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A fragment-based approach leading to the discovery of a novel binding site and the selective CK2 inhibitor CAM4066.
Bioorg. Med. Chem., 25, 2017
|
|
6UXF
 
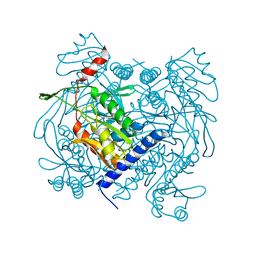 | | Structure of V. metoecus NucC, hexamer form | | Descriptor: | Vibrio meotecus sp. RC341 NucC | | Authors: | Ye, Q, Corbett, K.D. | | Deposit date: | 2019-11-07 | | Release date: | 2019-12-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structure and Mechanism of a Cyclic Trinucleotide-Activated Bacterial Endonuclease Mediating Bacteriophage Immunity.
Mol.Cell, 77, 2020
|
|
8CDU
 
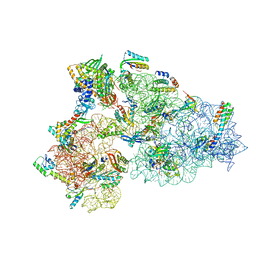 | | Rnase R bound to a 30S degradation intermediate (main state) | | Descriptor: | 16S rRNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Paternoga, H, Dimitrova-Paternoga, L, Wilson, D.N. | | Deposit date: | 2023-02-01 | | Release date: | 2023-12-20 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis of ribosomal 30S subunit degradation by RNase R.
Nature, 626, 2024
|
|
8CED
 
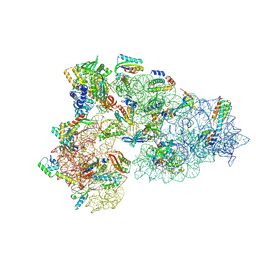 | | Rnase R bound to a 30S degradation intermediate (State I - head-turning) | | Descriptor: | 16S rRNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Paternoga, H, Dimitrova-Paternoga, L, Wilson, D.N. | | Deposit date: | 2023-02-01 | | Release date: | 2023-12-20 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4.15 Å) | | Cite: | Structural basis of ribosomal 30S subunit degradation by RNase R.
Nature, 626, 2024
|
|
5MIF
 
 | | Crystal structure of carboxyl esterase 2 (TmelEST2) from mycorrhizal fungus Tuber melanosporum | | Descriptor: | 'Carboxyl esterase 2, FRAGMENT OF TRITON X-100 | | Authors: | Zanotti, G, Vallese, F, Cavazzini, D, Ottonello, S. | | Deposit date: | 2016-11-28 | | Release date: | 2017-08-23 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.141 Å) | | Cite: | A family of archaea-like carboxylesterases preferentially expressed in the symbiotic phase of the mycorrhizal fungus Tuber melanosporum.
Sci Rep, 7, 2017
|
|
8CEC
 
 | | Rnase R bound to a 30S degradation intermediate (State I - head-turning) | | Descriptor: | 16S rRNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Paternoga, H, Dimitrova-Paternoga, L, Wilson, D.N. | | Deposit date: | 2023-02-01 | | Release date: | 2023-12-20 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.57 Å) | | Cite: | Structural basis of ribosomal 30S subunit degradation by RNase R.
Nature, 626, 2024
|
|
5CNJ
 
 | | mGlur2 with glutamate analog | | Descriptor: | (1R,2S,4R,5R,6R)-2-amino-4-(1H-1,2,4-triazol-3-ylsulfanyl)bicyclo[3.1.0]hexane-2,6-dicarboxylic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, Metabotropic glutamate receptor 2 | | Authors: | Monn, J.A, Clawson, D.K. | | Deposit date: | 2015-07-17 | | Release date: | 2015-09-09 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Synthesis and Pharmacological Characterization of C4-(Thiotriazolyl)-substituted-2-aminobicyclo[3.1.0]hexane-2,6-dicarboxylates. Identification of (1R,2S,4R,5R,6R)-2-Amino-4-(1H-1,2,4-triazol-3-ylsulfanyl)bicyclo[3.1.0]hexane-2,6-dicarboxylic Acid (LY2812223), a Highly Potent, Functionally Selective mGlu2 Receptor Agonist.
J.Med.Chem., 58, 2015
|
|
5JQ0
 
 | | Crystal structure of human carbonic anhydrase II in complex with Benzoxaborole at pH=8.7 | | Descriptor: | 1,1-dihydroxy-1,3-dihydro-2,1-benzoxaborol-1-ium, Carbonic anhydrase 2, ZINC ION | | Authors: | Alterio, V, Esposito, D, Di Fiore, A, De Simone, G. | | Deposit date: | 2016-05-04 | | Release date: | 2016-10-19 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Benzoxaborole as a new chemotype for carbonic anhydrase inhibition.
Chem.Commun.(Camb.), 52, 2016
|
|
5MO8
 
 | | Crystal Structure of CK2alpha with N-(3-(((2-chloro-[1,1'-biphenyl]-4-yl)methyl)amino)propyl)methanesulfonamide bound | | Descriptor: | 3-[[3-[3-[(3-chloranyl-4-phenyl-phenyl)methylamino]propylamino]-3-oxidanylidene-propanoyl]amino]benzoic acid, ACETATE ION, Casein kinase II subunit alpha, ... | | Authors: | Brear, P, De Fusco, C, Georgiou, K, Iegre, J, Sore, H, Hyvonen, M, Spring, D. | | Deposit date: | 2016-12-13 | | Release date: | 2017-05-24 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | A fragment-based approach leading to the discovery of a novel binding site and the selective CK2 inhibitor CAM4066.
Bioorg. Med. Chem., 25, 2017
|
|
4Y1D
 
 | |
8CDV
 
 | | Rnase R bound to a 30S degradation intermediate (state II) | | Descriptor: | 16S rRNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Paternoga, H, Dimitrova-Paternoga, L, Wilson, D.N. | | Deposit date: | 2023-02-01 | | Release date: | 2023-12-20 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4.73 Å) | | Cite: | Structural basis of ribosomal 30S subunit degradation by RNase R.
Nature, 626, 2024
|
|
