5N4B
 
 | | Prolyl oligopeptidase B from Galerina marginata bound to 25mer macrocyclization substrate - S577A mutant | | Descriptor: | Alpha-amanitin proprotein, Prolyl oligopeptidase | | Authors: | Czekster, C.M, McMahon, S.A, Ludewig, H, Naismith, J.H. | | Deposit date: | 2017-02-10 | | Release date: | 2017-11-01 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Characterization of a dual function macrocyclase enables design and use of efficient macrocyclization substrates.
Nat Commun, 8, 2017
|
|
5N4E
 
 | | Prolyl oligopeptidase B from Galerina marginata bound to 35mer hydrolysis and macrocyclization substrate - H698A mutant | | Descriptor: | Alpha-amanitin proprotein, GLYCEROL, Prolyl oligopeptidase | | Authors: | Czekster, C.M, McMahon, S.A, Ludewig, H, Naismith, J.H. | | Deposit date: | 2017-02-10 | | Release date: | 2017-11-01 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Characterization of a dual function macrocyclase enables design and use of efficient macrocyclization substrates.
Nat Commun, 8, 2017
|
|
5N4F
 
 | | Prolyl oligopeptidase B from Galerina marginata - apo protein | | Descriptor: | GLYCEROL, Prolyl oligopeptidase | | Authors: | Czekster, C.M, McMahon, S.A, Ludewig, H, Naismith, J.H. | | Deposit date: | 2017-02-10 | | Release date: | 2017-11-01 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Characterization of a dual function macrocyclase enables design and use of efficient macrocyclization substrates.
Nat Commun, 8, 2017
|
|
5N4D
 
 | | Prolyl oligopeptidase B from Galerina marginata bound to 25mer macrocyclization substrate - D661A mutant | | Descriptor: | Alpha-amanitin proprotein, GLYCEROL, Prolyl oligopeptidase | | Authors: | Czekster, C.M, McMahon, S.A, Ludewig, H, Naismith, J.H. | | Deposit date: | 2017-02-10 | | Release date: | 2017-11-01 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Characterization of a dual function macrocyclase enables design and use of efficient macrocyclization substrates.
Nat Commun, 8, 2017
|
|
5N4C
 
 | | Prolyl oligopeptidase B from Galerina marginata bound to 35mer hydrolysis and macrocyclization substrate - S577A mutant | | Descriptor: | Alpha-amanitin proprotein, GLYCEROL, Prolyl oligopeptidase | | Authors: | Czekster, C.M, McMahon, S.A, Ludewig, H, Naismith, J.H. | | Deposit date: | 2017-02-10 | | Release date: | 2017-11-01 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Characterization of a dual function macrocyclase enables design and use of efficient macrocyclization substrates.
Nat Commun, 8, 2017
|
|
8QC0
 
 | |
8PZ0
 
 | | Intracellular leucine aminopeptidase of Pseudomonas aeruginosa PA14. | | Descriptor: | 1,2-ETHANEDIOL, BICARBONATE ION, CHLORIDE ION, ... | | Authors: | Simpson, M.C, Czekster, C.M, Harding, C.J. | | Deposit date: | 2023-07-26 | | Release date: | 2023-11-15 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Unveiling the Catalytic Mechanism of a Processive Metalloaminopeptidase.
Biochemistry, 62, 2023
|
|
8PZM
 
 | | Intracellular leucine aminopeptidase of Pseudomonas aeruginosa PA14 bound to bestatin inhibitor and manganese | | Descriptor: | 1,2-ETHANEDIOL, 2-(3-AMINO-2-HYDROXY-4-PHENYL-BUTYRYLAMINO)-4-METHYL-PENTANOIC ACID, BICARBONATE ION, ... | | Authors: | Simpson, M.C, Czekster, C.M, Harding, C.J. | | Deposit date: | 2023-07-27 | | Release date: | 2023-11-15 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Unveiling the Catalytic Mechanism of a Processive Metalloaminopeptidase.
Biochemistry, 62, 2023
|
|
8PZY
 
 | | Intracellular leucine aminopeptidase of Pseudomonas aeruginosa PA14 - hexameric assembly with manganese bound | | Descriptor: | 1,2-ETHANEDIOL, 2-[3-[3-(2-hydroxyethoxy)propoxy]propoxy]ethanol, AMMONIUM ION, ... | | Authors: | Simpson, M.C, Czekster, C.M, Harding, C.J. | | Deposit date: | 2023-07-27 | | Release date: | 2023-11-15 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Unveiling the Catalytic Mechanism of a Processive Metalloaminopeptidase.
Biochemistry, 62, 2023
|
|
7O4O
 
 | | Structure of Staphylococcus aureus m1A22-tRNA methyltransferase in complex with S-adenosylhomocysteine | | Descriptor: | GLYCEROL, S-ADENOSYL-L-HOMOCYSTEINE, tRNA (Adenine(22)-N(1))-methyltransferase | | Authors: | Gloster, T.M, Czekster, C.M, da Silva, R.G. | | Deposit date: | 2021-04-06 | | Release date: | 2022-04-06 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Structure, dynamics, and molecular inhibition of the Staphylococcus aureus m 1 A22-tRNA methyltransferase TrmK.
J.Biol.Chem., 298, 2022
|
|
7O4N
 
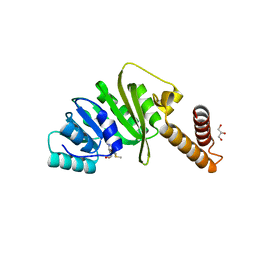 | | Structure of Staphylococcus aureus m1A22-tRNA methyltransferase in complex with S-adenosylmethionine | | Descriptor: | GLYCEROL, S-ADENOSYLMETHIONINE, tRNA (Adenine(22)-N(1))-methyltransferase | | Authors: | Gloster, T.M, Czekster, C.M, da Silva, R.G. | | Deposit date: | 2021-04-06 | | Release date: | 2022-04-20 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure, dynamics, and molecular inhibition of the Staphylococcus aureus m 1 A22-tRNA methyltransferase TrmK.
J.Biol.Chem., 298, 2022
|
|
7O4M
 
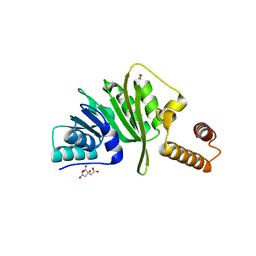 | | Structure of Staphylococcus aureus m1A22-tRNA methyltransferase | | Descriptor: | CITRIC ACID, GLYCEROL, tRNA (Adenine(22)-N(1))-methyltransferase | | Authors: | Gloster, T.M, Czekster, C.M, da Silva, R.G. | | Deposit date: | 2021-04-06 | | Release date: | 2022-04-27 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structure, dynamics, and molecular inhibition of the Staphylococcus aureus m 1 A22-tRNA methyltransferase TrmK.
J.Biol.Chem., 298, 2022
|
|
7AZU
 
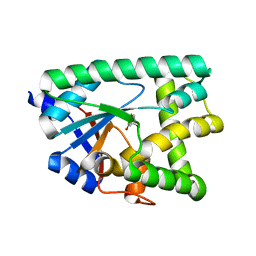 | |
6ZTU
 
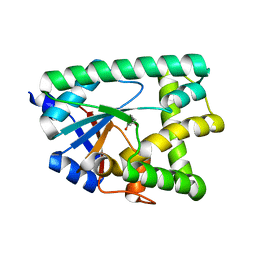 | |
6ZU3
 
 | |
8ACK
 
 | |
8ACR
 
 | |
8ACG
 
 | |
8AC7
 
 | |
8AC9
 
 | |
8PQR
 
 | | Nucleoside 2'deoxyribosyltransferase from Chroococcidiopsis thermalis PCC 7203 WT bound to DAD_Immucillin-H | | Descriptor: | 7-[[(3R,4R)-3-(hydroxymethyl)-4-oxidanyl-pyrrolidin-1-ium-1-yl]methyl]-3,5-dihydropyrrolo[3,2-d]pyrimidin-4-one, Nucleoside 2-deoxyribosyltransferase | | Authors: | Tang, P, Harding, C.J, Czekster, C.M. | | Deposit date: | 2023-07-11 | | Release date: | 2024-02-21 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.586 Å) | | Cite: | Snapshots of the Reaction Coordinate of a Thermophilic 2'-Deoxyribonucleoside/ribonucleoside Transferase.
Acs Catalysis, 14, 2024
|
|
8PQT
 
 | |
8PQS
 
 | |
8RH3
 
 | | Nucleoside 2'deoxyribosyltransferase from Chroococcidiopsis thermalis PCC 7203 WT bound to Gemcitabine | | Descriptor: | (2~{R},3~{R})-4,4-bis(fluoranyl)-2-(hydroxymethyl)oxolan-3-ol, Nucleoside 2-deoxyribosyltransferase | | Authors: | Tang, P, Harding, C.J, Czekster, C.M. | | Deposit date: | 2023-12-14 | | Release date: | 2024-02-21 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Snapshots of the Reaction Coordinate of a Thermophilic 2'-Deoxyribonucleoside/ribonucleoside Transferase.
Acs Catalysis, 14, 2024
|
|
7QAT
 
 | |
