2FC2
 
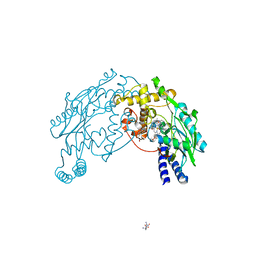 | |
2I7O
 
 | | Structure of Re(4,7-dimethyl-phen)(Thr124His)(Lys122Trp)(His83Gln)AzCu(II), a Rhenium modified Azurin mutant | | Descriptor: | (1,10 PHENANTHROLINE)-(TRI-CARBON MONOXIDE) RHENIUM (I), Azurin, COPPER (II) ION | | Authors: | Sudhamsu, J, Crane, B.R. | | Deposit date: | 2006-08-31 | | Release date: | 2007-08-14 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Tryptophan-accelerated electron flow through proteins.
Science, 320, 2008
|
|
2FNW
 
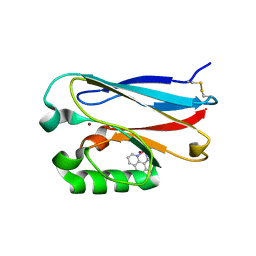 | | Pseudomonas aeruginosa E2Q/H83Q/M109H-azurin RE(PHEN)(CO)3 | | Descriptor: | (1,10 PHENANTHROLINE)-(TRI-CARBON MONOXIDE) RHENIUM (I), Azurin, COPPER (II) ION | | Authors: | Gradinaru, C, Crane, B.R. | | Deposit date: | 2006-01-11 | | Release date: | 2006-10-03 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Excited-state dynamics of structurally characterized [ReI(CO)3(phen)(HisX)]+ (X = 83, 109) Pseudomonas aeruginosa azurins in aqueous solution.
J.Am.Chem.Soc., 128, 2006
|
|
2NOX
 
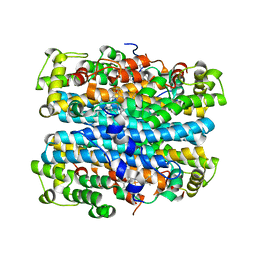 | | Crystal structure of tryptophan 2,3-dioxygenase from Ralstonia metallidurans | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, Tryptophan 2,3-dioxygenase | | Authors: | Zhang, Y, Kang, S.A, Mukherjee, T, Bale, S, Crane, B.R, Begley, T.P, Ealick, S.E. | | Deposit date: | 2006-10-26 | | Release date: | 2006-12-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure and mechanism of tryptophan 2,3-dioxygenase, a heme enzyme involved in tryptophan catabolism and in quinolinate biosynthesis.
Biochemistry, 46, 2007
|
|
2PD8
 
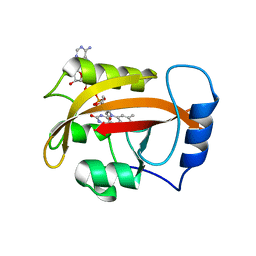 | |
2PD7
 
 | |
2I7S
 
 | | Crystal structure of Re(phen)(CO)3 (Thr124His)(His83Gln) Azurin Cu(II) from Pseudomonas aeruginosa | | Descriptor: | (1,10 PHENANTHROLINE)-(TRI-CARBON MONOXIDE) RHENIUM (I), Azurin, COBALT TETRAAMMINE ION, ... | | Authors: | Gradinaru, C, Crane, B.R. | | Deposit date: | 2006-08-31 | | Release date: | 2006-12-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Relaxation dynamics of Pseudomonas aeruginosa Re(I)(CO)3(alpha-diimine)(HisX)+ (X = 83, 107, 109, 124, 126)Cu(II) azurins.
J.Am.Chem.Soc., 131, 2009
|
|
2PDR
 
 | |
2CH7
 
 | |
2CH4
 
 | |
2B11
 
 | |
2B10
 
 | |
2BCN
 
 | |
2AMO
 
 | |
2AN0
 
 | |
2B0Z
 
 | |
2F9Z
 
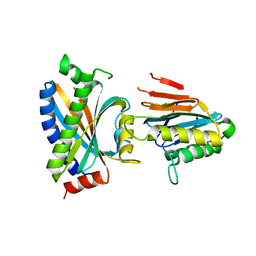 | | Complex between the chemotaxis deamidase CheD and the chemotaxis phosphatase CheC from Thermotoga maritima | | Descriptor: | PROTEIN (chemotaxis methylation protein), chemotaxis protein CheC | | Authors: | Chao, X, Park, S.Y, Bilwes, A.M, Crane, B.R. | | Deposit date: | 2005-12-06 | | Release date: | 2006-06-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A receptor-modifying deamidase in complex with a signaling phosphatase reveals reciprocal regulation.
Cell(Cambridge,Mass.), 124, 2006
|
|
2FC1
 
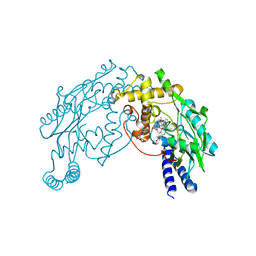 | | Heme NO Complex in NOS | | Descriptor: | 7,8-DIHYDROBIOPTERIN, ARGININE, NITRIC OXIDE, ... | | Authors: | Pant, K, Crane, B.R. | | Deposit date: | 2005-12-10 | | Release date: | 2006-04-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Nitrosyl-Heme Structures of Bacillus subtilis Nitric Oxide Synthase Have Implications for Understanding Substrate Oxidation.
Biochemistry, 45, 2006
|
|
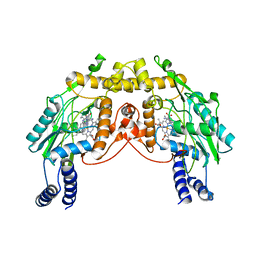
2FLQ
 
 | |
2FBZ
 
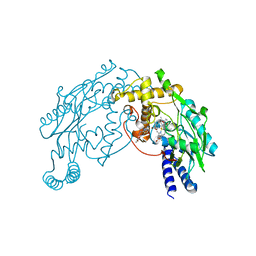 | | Heme-No complex in a bacterial Nitric Oxide Synthase | | Descriptor: | 2-AMINO-6-(1,2-DIHYDROXY-PROPYL)-7,8-DIHYDRO-6H-PTERIDIN-4-ONE, N-OMEGA-HYDROXY-L-ARGININE, NITRIC OXIDE, ... | | Authors: | Pant, K, Crane, B.R. | | Deposit date: | 2005-12-10 | | Release date: | 2006-08-29 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Nitrosyl-heme structures of Bacillus subtilis nitric oxide synthase have implications for understanding substrate oxidation.
Biochemistry, 45, 2006
|
|
3E67
 
 | | Murine inos dimer with inhibitor 4-MAP bound | | Descriptor: | 4-METHYLPYRIDIN-2-AMINE, 5,6,7,8-TETRAHYDROBIOPTERIN, Nitric oxide synthase, ... | | Authors: | Garcin, E.D, Arvai, A.S, Rosenfeld, R.J, Kroeger, M.D, Crane, B.R, Andersson, G, Andrews, G, Hamley, P.J, Mallinder, P.R, Nicholls, D.J, St-Gallay, S.A, Tinker, A.C, Gensmantel, N.P, Mete, A, Cheshire, D.R, Connolly, S, Stueh, D.J, Aberg, A, Wallace, A.V, Tainer, J.A, Getzoff, E.D. | | Deposit date: | 2008-08-14 | | Release date: | 2008-10-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Anchored plasticity opens doors for selective inhibitor design in nitric oxide synthase.
Nat.Chem.Biol., 4, 2008
|
|
3E6T
 
 | | Structure of murine INOS oxygenase domain with inhibitor AR-C118901 | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, 5-(4'-AMINO-1'-ETHYL-5',8'-DIFLUORO-1'H-SPIRO[PIPERIDINE-4,2'-QUINAZOLINE]-1-YLCARBONYL)PICOLINONITRILE, Nitric oxide synthase, ... | | Authors: | Garcin, E.D, Arvai, A.S, Rosenfeld, R.J, Kroeger, M.D, Crane, B.R, Andersson, G, Andrews, G, Hamley, P.J, Mallinder, P.R, Nicholls, D.J, St-Gallay, S.A, Tinker, A.C, Gensmantel, N.P, Mete, A, Cheshire, D.R, Connolly, S, Stueh, D.J, Aberg, A, Wallace, A.V, Tainer, J.A, Getzoff, E.D. | | Deposit date: | 2008-08-15 | | Release date: | 2008-10-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Anchored plasticity opens doors for selective inhibitor design in nitric oxide synthase.
Nat.Chem.Biol., 4, 2008
|
|
3E68
 
 | | Structure of murine INOS oxygenase domain with inhibitor AR-C130232 | | Descriptor: | 1,2-ETHANEDIOL, 5,6,7,8-TETRAHYDROBIOPTERIN, N-[2-(6-AMINO-4-METHYLPYRIDIN-2-YL)ETHYL]-4-CYANOBENZAMIDE, ... | | Authors: | Garcin, E.D, Arvai, A.S, Rosenfeld, R.J, Kroeger, M.D, Crane, B.R, Andersson, G, Andrews, G, Hamley, P.J, Mallinder, P.R, Nicholls, D.J, St-Gallay, S.A, Tinker, A.C, Gensmantel, N.P, Mete, A, Cheshire, D.R, Connolly, S, Stueh, D.J, Aberg, A, Wallace, A.V, Tainer, J.A, Getzoff, E.D. | | Deposit date: | 2008-08-14 | | Release date: | 2008-10-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Anchored plasticity opens doors for selective inhibitor design in nitric oxide synthase.
Nat.Chem.Biol., 4, 2008
|
|
3E7M
 
 | | Structure of murine iNOS oxygenase domain with inhibitor AR-C95791 | | Descriptor: | 1,2-ETHANEDIOL, 5,6,7,8-TETRAHYDROBIOPTERIN, ETHYL 4-[(4-METHYLPYRIDIN-2-YL)AMINO]PIPERIDINE-1-CARBOXYLATE, ... | | Authors: | Garcin, E.D, Arvai, A.S, Rosenfeld, R.J, Kroeger, M.D, Crane, B.R, Andersson, G, Andrews, G, Hamley, P.J, Mallinder, P.R, Nicholls, D.J, St-Gallay, S.A, Tinker, A.C, Gensmantel, N.P, Mete, A, Cheshire, D.R, Connolly, S, Stuehr, D.J, Aberg, A, Wallace, A.V, Tainer, J.A, Getzoff, E.D. | | Deposit date: | 2008-08-18 | | Release date: | 2008-10-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Anchored plasticity opens doors for selective inhibitor design in nitric oxide synthase.
Nat.Chem.Biol., 4, 2008
|
|
3EC1
 
 | |
