4JU2
 
 | | Crystal structure of hcv ns5b polymerase in complex with compound 12 | | 分子名称: | 6-[3-(pyridin-2-yl)-2-(trifluoromethyl)phenoxy]-1-(2,4,6-trifluorobenzyl)quinazolin-4(1H)-one, GLYCEROL, Genome polyprotein, ... | | 著者 | Coulombe, R. | | 登録日 | 2013-03-24 | | 公開日 | 2013-07-03 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Molecular Dynamics Simulations and Structure-Based Rational Design Lead to Allosteric HCV NS5B Polymerase Thumb Pocket 2 Inhibitor with Picomolar Cellular Replicon Potency.
J.Med.Chem., 57, 2014
|
|
4JTY
 
 | | Crystal structure of HCV NS5B polymerase with COMPOUND 2 | | 分子名称: | 3-{[4-oxo-1-(2,4,6-trifluorobenzyl)-1,4-dihydroquinazolin-6-yl]oxy}-2-(trifluoromethyl)benzamide, GLYCEROL, Genome polyprotein, ... | | 著者 | Coulombe, R. | | 登録日 | 2013-03-24 | | 公開日 | 2013-07-03 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Molecular Dynamics Simulations and Structure-Based Rational Design Lead to Allosteric HCV NS5B Polymerase Thumb Pocket 2 Inhibitor with Picomolar Cellular Replicon Potency.
J.Med.Chem., 57, 2014
|
|
4JU4
 
 | | Crystal structure of hcv ns5b polymerase in complex with compound 22 | | 分子名称: | 2-{[(4-bromo-2-fluorophenyl)sulfonyl]amino}-5-phenoxybenzoic acid, Genome polyprotein, MAGNESIUM ION | | 著者 | Coulombe, R. | | 登録日 | 2013-03-24 | | 公開日 | 2013-11-27 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Anthranilic acid-based Thumb Pocket 2 HCV NS5B polymerase inhibitors with sub-micromolar potency in the cell-based replicon assay.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
1MIR
 
 | | RAT PROCATHEPSIN B | | 分子名称: | PROCATHEPSIN B | | 著者 | Cygler, M, Sivaraman, J, Grochulski, P, Coulombe, R, Storer, A.C, Mort, J.S. | | 登録日 | 1996-01-12 | | 公開日 | 1997-01-11 | | 最終更新日 | 2021-11-03 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Structure of rat procathepsin B: model for inhibition of cysteine protease activity by the proregion.
Structure, 4, 1996
|
|
1R6N
 
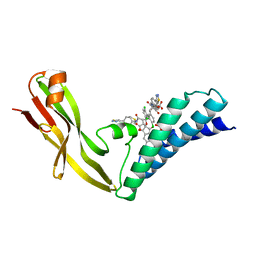 | | HPV11 E2 TAD complex crystal structure | | 分子名称: | 2-METHYL-PROPIONIC ACID, DIMETHYL SULFOXIDE, HPV11 REGULATORY PROTEIN E2, ... | | 著者 | Wang, Y, Coulombe, R. | | 登録日 | 2003-10-15 | | 公開日 | 2004-02-24 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Crystal Structure of the E2 Transactivation Domain of Human
Papillomavirus Type 11 Bound to a Protein Interaction Inhibitor
J.Biol.Chem., 279, 2004
|
|
1R6K
 
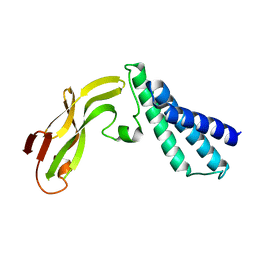 | | HPV11 E2 TAD crystal structure | | 分子名称: | HPV11 REGULATORY PROTEIN E2 | | 著者 | Wang, Y, Coulombe, R. | | 登録日 | 2003-10-15 | | 公開日 | 2004-02-24 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Crystal Structure of the E2 Transactivation Domain of Human Papillomavirus Type 11 Bound to a Protein Interaction Inhibitor
J.Biol.Chem., 279, 2004
|
|
1F8R
 
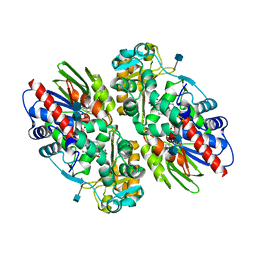 | | CRYSTAL STRUCTURE OF L-AMINO ACID OXIDASE FROM CALLOSELASMA RHODOSTOMA COMPLEXED WITH CITRATE | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, CITRIC ACID, ... | | 著者 | Pawelek, P.D, Cheah, J, Coulombe, R, Macheroux, P, Ghisla, S, Vrielink, A. | | 登録日 | 2000-07-04 | | 公開日 | 2000-08-24 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | The structure of L-amino acid oxidase reveals the substrate trajectory into an enantiomerically conserved active site.
EMBO J., 19, 2000
|
|
1F8S
 
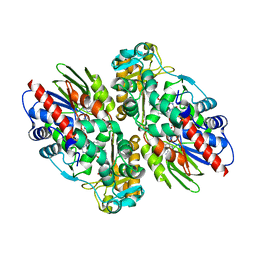 | | CRYSTAL STRUCTURE OF L-AMINO ACID OXIDASE FROM CALLOSELASMA RHODOSTOMA, COMPLEXED WITH THREE MOLECULES OF O-AMINOBENZOATE. | | 分子名称: | 2-AMINOBENZOIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, FLAVIN-ADENINE DINUCLEOTIDE, ... | | 著者 | Pawelek, P.D, Cheah, J, Coulombe, R, Macheroux, P, Ghisla, S, Vrielink, A. | | 登録日 | 2000-07-04 | | 公開日 | 2000-08-24 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | The structure of L-amino acid oxidase reveals the substrate trajectory into an enantiomerically conserved active site.
EMBO J., 19, 2000
|
|
1CS8
 
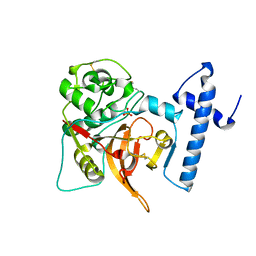 | |
8T5G
 
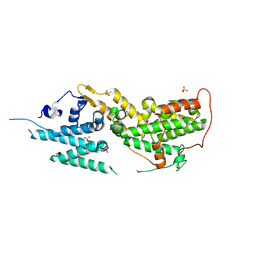 | | SOS2 co-crystal structure with fragment bound (compound 12) | | 分子名称: | DIMETHYL SULFOXIDE, SULFATE ION, Son of sevenless homolog 2, ... | | 著者 | Gunn, R.J, Lawson, J.D, Ivetac, A, Ulaganathan, T, Coulombe, R, Fethiere, J. | | 登録日 | 2023-06-13 | | 公開日 | 2024-01-10 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.92 Å) | | 主引用文献 | Discovery of Five SOS2 Fragment Hits with Binding Modes Determined by SOS2 X-Ray Cocrystallography.
J.Med.Chem., 67, 2024
|
|
8T5M
 
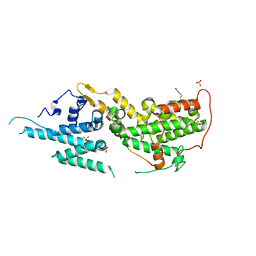 | | SOS2 crystal structure with fragment bound (compound 14) | | 分子名称: | 1,2-ETHANEDIOL, 4-[(1R,2S)-1-hydroxy-2-{[2-(4-hydroxyphenyl)ethyl]amino}propyl]phenol, SULFATE ION, ... | | 著者 | Gunn, R.J, Lawson, J.D, Ivetac, A, Ulaganathan, T, Coulombe, R, Fethiere, J. | | 登録日 | 2023-06-14 | | 公開日 | 2024-01-10 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.79 Å) | | 主引用文献 | Discovery of Five SOS2 Fragment Hits with Binding Modes Determined by SOS2 X-Ray Cocrystallography.
J.Med.Chem., 67, 2024
|
|
8T5R
 
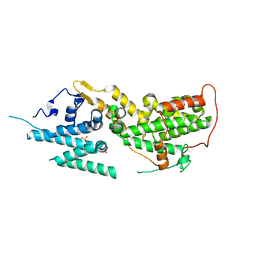 | | SOS2 crystal structure with fragment bound (compound 13) | | 分子名称: | 4-(aminomethyl)benzene-1-sulfonamide, SULFATE ION, Son of sevenless homolog 2 | | 著者 | Gunn, R.J, Lawson, J.D, Ivetac, A, Ulaganathan, T, Coulombe, R, Fethiere, J. | | 登録日 | 2023-06-14 | | 公開日 | 2024-01-10 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.12 Å) | | 主引用文献 | Discovery of Five SOS2 Fragment Hits with Binding Modes Determined by SOS2 X-Ray Cocrystallography.
J.Med.Chem., 67, 2024
|
|
1DOS
 
 | | STRUCTURE OF FRUCTOSE-BISPHOSPHATE ALDOLASE | | 分子名称: | ALDOLASE CLASS II, AMMONIUM ION, ZINC ION | | 著者 | Blom, N, Tetreault, S, Coulombe, R, Sygusch, J. | | 登録日 | 1996-06-24 | | 公開日 | 1997-07-07 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.67 Å) | | 主引用文献 | Novel active site in Escherichia coli fructose 1,6-bisphosphate aldolase.
Nat.Struct.Biol., 3, 1996
|
|
1EZ0
 
 | | CRYSTAL STRUCTURE OF THE NADP+ DEPENDENT ALDEHYDE DEHYDROGENASE FROM VIBRIO HARVEYI. | | 分子名称: | ALDEHYDE DEHYDROGENASE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | 著者 | Ahvazi, B, Coulombe, R, Delarge, M, Vedadi, M, Zhang, L, Meighen, E, Vrielink, A. | | 登録日 | 2000-05-09 | | 公開日 | 2000-05-24 | | 最終更新日 | 2023-08-09 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Crystal structure of the NADP+-dependent aldehyde dehydrogenase from Vibrio harveyi: structural implications for cofactor specificity and affinity.
Biochem.J., 349, 2000
|
|
1EYY
 
 | | CRYSTAL STRUCTURE OF THE NADP+ DEPENDENT ALDEHYDE DEHYDROGENASE FROM VIBRIO HARVEYI. | | 分子名称: | ALDEHYDE DEHYDROGENASE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | 著者 | Ahvazi, B, Coulombe, R, Delarge, M, Vedadi, M, Zhang, L, Meighen, E, Vrielink, A. | | 登録日 | 2000-05-09 | | 公開日 | 2000-05-24 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Crystal structure of the NADP+-dependent aldehyde dehydrogenase from Vibrio harveyi: structural implications for cofactor specificity and affinity.
Biochem.J., 349, 2000
|
|
