7Z13
 
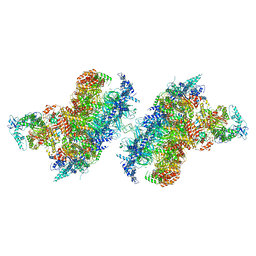 | | S. cerevisiae CMGE dimer nucleating origin DNA melting | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Cell division control protein 45, ... | | Authors: | Lewis, J.S, Sousa, J.S, Costa, A. | | Deposit date: | 2022-02-24 | | Release date: | 2022-06-15 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Mechanism of replication origin melting nucleated by CMG helicase assembly.
Nature, 606, 2022
|
|
6HV9
 
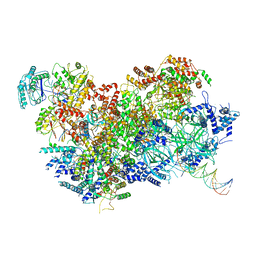 | | S. cerevisiae CMG-Pol epsilon-DNA | | Descriptor: | Cell division control protein 45, DNA (5'-D(*GP*CP*AP*GP*CP*CP*AP*CP*GP*CP*TP*GP*GP*CP*CP*GP*TP*TP*TP*TP*A)-3'), DNA (5'-D(P*TP*AP*AP*AP*AP*CP*GP*GP*CP*CP*AP*GP*CP*GP*TP*GP*GP*CP*TP*GP*C)-3'), ... | | Authors: | Abid Ali, F, Purkiss, A.G, Cheung, A, Costa, A. | | Deposit date: | 2018-10-10 | | Release date: | 2018-12-12 | | Last modified: | 2023-07-05 | | Method: | ELECTRON MICROSCOPY (4.98 Å) | | Cite: | Structure of DNA-CMG-Pol epsilon elucidates the roles of the non-catalytic polymerase modules in the eukaryotic replisome.
Nat Commun, 9, 2018
|
|
6HV8
 
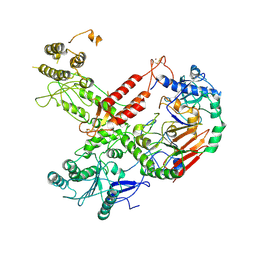 | | Cryo-EM structure of S. cerevisiae Polymerase epsilon deltacat mutant | | Descriptor: | DNA polymerase epsilon catalytic subunit A, DNA polymerase epsilon subunit B, ZINC ION | | Authors: | Goswami, P, Purkiss, A, Cheung, A, Costa, A. | | Deposit date: | 2018-10-10 | | Release date: | 2018-12-12 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Structure of DNA-CMG-Pol epsilon elucidates the roles of the non-catalytic polymerase modules in the eukaryotic replisome.
Nat Commun, 9, 2018
|
|
8Q6P
 
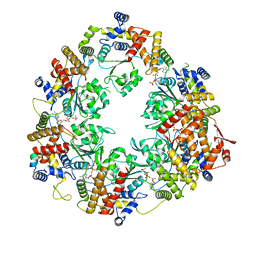 | | X. laevis CMG dimer bound to dimeric DONSON - MCM ATPase | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, DNA replication licensing factor mcm2, DNA replication licensing factor mcm4-B, ... | | Authors: | Butryn, A, Cvetkovic, M.A, Costa, A. | | Deposit date: | 2023-08-14 | | Release date: | 2023-10-18 | | Last modified: | 2023-11-29 | | Method: | ELECTRON MICROSCOPY (3.53 Å) | | Cite: | The structural mechanism of dimeric DONSON in replicative helicase activation.
Mol.Cell, 83, 2023
|
|
8Q6O
 
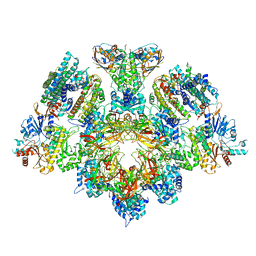 | | X. laevis CMG dimer bound to dimeric DONSON - without ATPase | | Descriptor: | Cell division control protein 45 homolog, DNA replication complex GINS protein PSF1, DNA replication complex GINS protein PSF2, ... | | Authors: | Butryn, A, Cvetkovic, M.A, Costa, A. | | Deposit date: | 2023-08-14 | | Release date: | 2023-10-18 | | Last modified: | 2023-11-29 | | Method: | ELECTRON MICROSCOPY (3.14 Å) | | Cite: | The structural mechanism of dimeric DONSON in replicative helicase activation.
Mol.Cell, 83, 2023
|
|
7ZPP
 
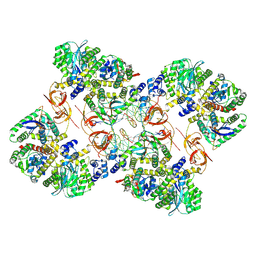 | | Cryo-EM structure of the MVV CSC intasome at 4.5A resolution | | Descriptor: | Integrase, vDNA, non-transferred strand, ... | | Authors: | Ballandras-Colas, A, Maskell, D, Pye, V.E, Locke, J, Swuec, S, Kotecha, A, Costa, A, Cherepanov, P. | | Deposit date: | 2022-04-28 | | Release date: | 2022-05-11 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | A supramolecular assembly mediates lentiviral DNA integration
Science, 355, 2017
|
|
5M0R
 
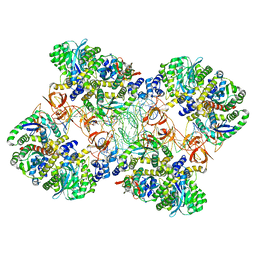 | | Cryo-EM reconstruction of the maedi-visna virus (MVV) strand transfer complex | | Descriptor: | integrase, tDNA, vDNA, ... | | Authors: | Pye, V.E, Ballandras-Colas, A, Maskell, D, Locke, J, Kotecha, A, Costa, A, Cherepanov, P. | | Deposit date: | 2016-10-05 | | Release date: | 2017-01-18 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (8.2 Å) | | Cite: | A supramolecular assembly mediates lentiviral DNA integration.
Science, 355, 2017
|
|
6F0L
 
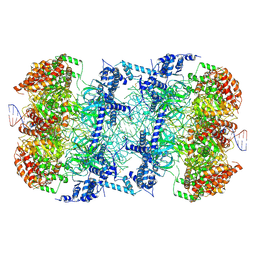 | | S. cerevisiae MCM double hexamer bound to duplex DNA | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA (62-MER), DNA replication licensing factor MCM2, ... | | Authors: | Abid Ali, F, Pye, V.E, Douglas, M.E, Locke, J, Nans, A, Diffley, J.F.X, Costa, A. | | Deposit date: | 2017-11-20 | | Release date: | 2017-12-06 | | Last modified: | 2019-12-11 | | Method: | ELECTRON MICROSCOPY (4.77 Å) | | Cite: | Cryo-EM structure of a licensed DNA replication origin.
Nat Commun, 8, 2017
|
|
8S0B
 
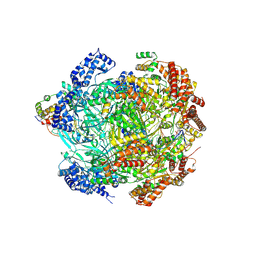 | | H. sapiens MCM bound to double stranded DNA and ORC6 as part of the MCM-ORC complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA (45-mer), DNA replication licensing factor MCM2, ... | | Authors: | Greiwe, J.F, Weissmann, F, Diffley, J.F.X, Costa, A. | | Deposit date: | 2024-02-13 | | Release date: | 2024-10-02 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | MCM Double Hexamer Loading Visualised with Human Proteins
Nature, 2024
|
|
8S0F
 
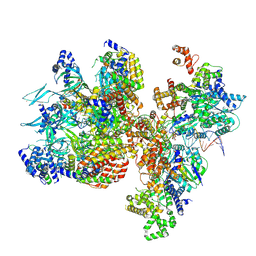 | | H. sapiens OC1M bound to double stranded DNA | | Descriptor: | DNA (39-mer), DNA replication factor Cdt1, DNA replication licensing factor MCM2, ... | | Authors: | Greiwe, J.F, Weissmann, F, Diffley, J.F.X, Costa, A. | | Deposit date: | 2024-02-13 | | Release date: | 2024-10-02 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | MCM Double Hexamer Loading Visualised with Human Proteins
Nature, 2024
|
|
8S0D
 
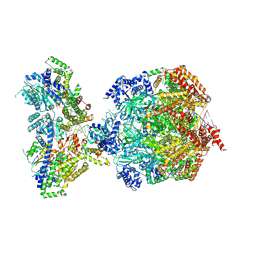 | | H. sapiens MCM bound to double stranded DNA and ORC1-6 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA (58-mer), DNA replication licensing factor MCM2, ... | | Authors: | Greiwe, J.F, Weissmann, F, Diffley, J.F.X, Costa, A. | | Deposit date: | 2024-02-13 | | Release date: | 2024-10-02 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | MCM Double Hexamer Loading Visualised with Human Proteins
Nature, 2024
|
|
8S0E
 
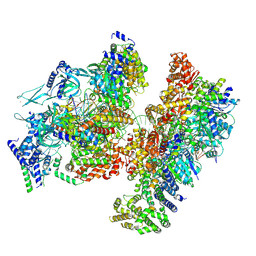 | | H. sapiens OCCM bound to double stranded DNA | | Descriptor: | Cell division control protein 6 homolog, DNA (39-mer), DNA replication factor Cdt1, ... | | Authors: | Greiwe, J.F, Weissmann, F, Diffley, J.F.X, Costa, A. | | Deposit date: | 2024-02-13 | | Release date: | 2024-10-02 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | MCM Double Hexamer Loading Visualised with Human Proteins
Nature, 2024
|
|
8S0A
 
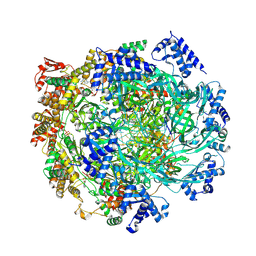 | | H. sapiens MCM2-7 hexamer bound to double stranded DNA | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, DNA (22-mer), ... | | Authors: | Greiwe, J.F, Weissmann, F, Diffley, J.F.X, Costa, A. | | Deposit date: | 2024-02-13 | | Release date: | 2024-10-02 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | MCM Double Hexamer Loading Visualised with Human Proteins
Nature, 2024
|
|
8S09
 
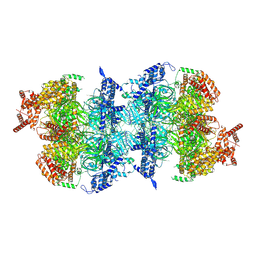 | | H. sapiens MCM2-7 double hexamer bound to double stranded DNA | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, DNA (45-mer), ... | | Authors: | Greiwe, J.F, Weissmann, F, Diffley, J.F.X, Costa, A. | | Deposit date: | 2024-02-13 | | Release date: | 2024-10-02 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | MCM Double Hexamer Loading Visualised with Human Proteins
Nature, 2024
|
|
8S0C
 
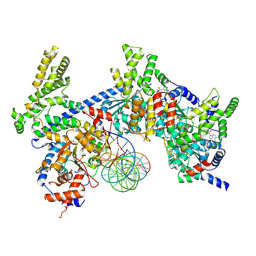 | | H. sapiens ORC1-5 bound to double stranded DNA as part of the MCM-ORC complex | | Descriptor: | DNA (26-mer), Isoform 2 of Origin recognition complex subunit 3, MAGNESIUM ION, ... | | Authors: | Greiwe, J.F, Weissmann, F, Diffley, J.F.X, Costa, A. | | Deposit date: | 2024-02-13 | | Release date: | 2024-10-02 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | MCM Double Hexamer Loading Visualised with Human Proteins
Nature, 2024
|
|
6RNY
 
 | | PFV intasome - nucleosome strand transfer complex | | Descriptor: | DNA (108-MER), DNA (128-MER), DNA (33-MER), ... | | Authors: | Pye, V.E, Renault, L, Maskell, D.P, Cherepanov, P, Costa, A. | | Deposit date: | 2019-05-09 | | Release date: | 2019-09-25 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Retroviral integration into nucleosomes through DNA looping and sliding along the histone octamer.
Nat Commun, 10, 2019
|
|
6YUF
 
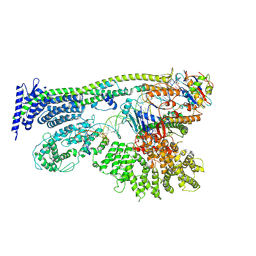 | | Cohesin complex with loader gripping DNA | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, Cohesin subunit rad21, ... | | Authors: | Higashi, T.L, Eickhoff, P, Sousa, J.S, Costa, A, Uhlmann, F. | | Deposit date: | 2020-04-27 | | Release date: | 2020-08-19 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.94 Å) | | Cite: | A Structure-Based Mechanism for DNA Entry into the Cohesin Ring.
Mol.Cell, 79, 2020
|
|
4BXO
 
 | | Architecture and DNA recognition elements of the Fanconi anemia FANCM- FAAP24 complex | | Descriptor: | 5'-D(*GP*AP*TP*GP*AP*TP*GP*CP*TP*GP*CP)-3', 5'-D(*TP*CP*AP*GP*CP*AP*TP*CP*AP*TP*CP)-3', CALCIUM ION, ... | | Authors: | Coulthard, R, Deans, A, Swuec, P, Bowles, M, Purkiss, A, Costa, A, West, S, McDonald, N. | | Deposit date: | 2013-07-15 | | Release date: | 2013-08-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Architecture and DNA Recognition Elements of the Fanconi Anemia Fancm-Faap24 Complex.
Structure, 21, 2013
|
|
7QHS
 
 | | S. cerevisiae CMGE nucleating origin DNA melting | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Cell division control protein 45, ... | | Authors: | Lewis, J.S, Sousa, J.S, Costa, A. | | Deposit date: | 2021-12-14 | | Release date: | 2022-06-15 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Mechanism of replication origin melting nucleated by CMG helicase assembly.
Nature, 606, 2022
|
|
4AU8
 
 | | Crystal structure of compound 4a in complex with cdk5, showing an unusual binding mode to the hinge region via a water molecule | | Descriptor: | 4-(1,3-benzothiazol-2-yl)thiophene-2-sulfonamide, CYCLIN-DEPENDENT KINASE 5, IMIDAZOLE, ... | | Authors: | Malmstrom, J, Viklund, J, Slivo, C, Costa, A, Maudet, M, Sandelin, C, Hiller, G, Olsson, L.L, Aagaard, A, Geschwindner, S, Xue, Y, Vasange, M. | | Deposit date: | 2012-05-14 | | Release date: | 2013-03-27 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Synthesis and Structure-Activity Relationship of 4-(1,3-Benzothiazol-2-Yl)-Thiophene-2-Sulfonamides as Cyclin-Dependent Kinase 5 (Cdk5)/P25 Inhibitors.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
6R89
 
 | | Structure of Arabidopsis thaliana GLR3.3 ligand-binding domain in complex with L-cysteine | | Descriptor: | CHLORIDE ION, CYSTEINE, GLYCEROL, ... | | Authors: | Alfieri, A, Pederzoli, R, Costa, A. | | Deposit date: | 2019-04-01 | | Release date: | 2020-01-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The structural bases for agonist diversity in anArabidopsis thalianaglutamate receptor-like channel.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6R88
 
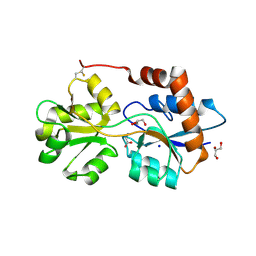 | | Structure of Arabidopsis thaliana GLR3.3 ligand-binding domain in complex with glycine | | Descriptor: | CHLORIDE ION, GLYCEROL, GLYCINE, ... | | Authors: | Alfieri, A, Pederzoli, R, Costa, A. | | Deposit date: | 2019-04-01 | | Release date: | 2020-01-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The structural bases for agonist diversity in anArabidopsis thalianaglutamate receptor-like channel.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6R85
 
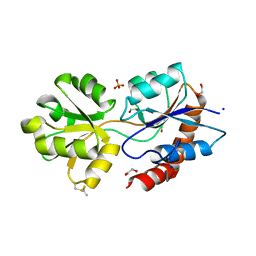 | | Structure of Arabidopsis thaliana GLR3.3 ligand-binding domain in complex with L-glutamate | | Descriptor: | 1,2-ETHANEDIOL, GLUTAMIC ACID, Glutamate receptor 3.3,Glutamate receptor 3.3, ... | | Authors: | Alfieri, A, Pederzoli, R, Costa, A. | | Deposit date: | 2019-03-31 | | Release date: | 2020-01-01 | | Last modified: | 2020-01-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The structural bases for agonist diversity in anArabidopsis thalianaglutamate receptor-like channel.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6R8A
 
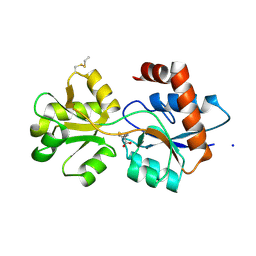 | | Structure of Arabidopsis thaliana GLR3.3 ligand-binding domain in complex with L-methionine | | Descriptor: | Glutamate receptor 3.3,Glutamate receptor 3.3, METHIONINE, SODIUM ION, ... | | Authors: | Alfieri, A, Pederzoli, R, Costa, A. | | Deposit date: | 2019-04-01 | | Release date: | 2020-01-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | The structural bases for agonist diversity in anArabidopsis thalianaglutamate receptor-like channel.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6R0C
 
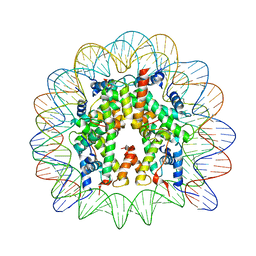 | | Human-D02 Nucleosome Core Particle with biotin-streptavidin label | | Descriptor: | DNA (142-MER), Histone H2A type 1, Histone H2B type 1-C/E/F/G/I, ... | | Authors: | Pye, V.E, Wilson, M.D, Cherepanov, P, Costa, A. | | Deposit date: | 2019-03-12 | | Release date: | 2019-09-25 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Retroviral integration into nucleosomes through DNA looping and sliding along the histone octamer.
Nat Commun, 10, 2019
|
|
