4TZJ
 
 | |
4TZM
 
 | |
4TZN
 
 | | Structure of HTP-2 bound to HTP-3 motif-6 | | Descriptor: | Protein HTP-2, Protein HTP-3 | | Authors: | Rosenberg, S.C, Corbett, K.D. | | Deposit date: | 2014-07-10 | | Release date: | 2014-11-19 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.115 Å) | | Cite: | The Chromosome Axis Controls Meiotic Events through a Hierarchical Assembly of HORMA Domain Proteins.
Dev.Cell, 31, 2014
|
|
6UXF
 
 | | Structure of V. metoecus NucC, hexamer form | | Descriptor: | Vibrio meotecus sp. RC341 NucC | | Authors: | Ye, Q, Corbett, K.D. | | Deposit date: | 2019-11-07 | | Release date: | 2019-12-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structure and Mechanism of a Cyclic Trinucleotide-Activated Bacterial Endonuclease Mediating Bacteriophage Immunity.
Mol.Cell, 77, 2020
|
|
6UXG
 
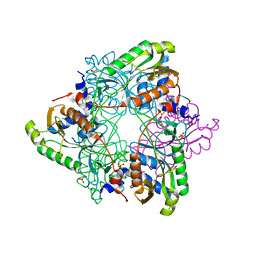 | | Structure of V. metoecus NucC, trimer form | | Descriptor: | SULFATE ION, Vibrio metoecus sp. RC341 NucC | | Authors: | Ye, Q, Corbett, K.D. | | Deposit date: | 2019-11-07 | | Release date: | 2019-12-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure and Mechanism of a Cyclic Trinucleotide-Activated Bacterial Endonuclease Mediating Bacteriophage Immunity.
Mol.Cell, 77, 2020
|
|
6P8O
 
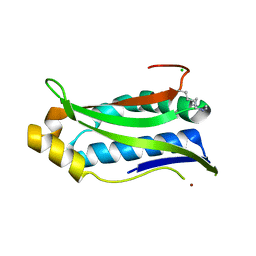 | | Structure of P. aeruginosa ATCC27853 HORMA2-deltaC | | Descriptor: | CHLORIDE ION, HORMA domain containing protein, NICKEL (II) ION | | Authors: | Ye, Q, Corbett, K.D, Lau, R.K. | | Deposit date: | 2019-06-07 | | Release date: | 2019-12-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.604 Å) | | Cite: | HORMA Domain Proteins and a Trip13-like ATPase Regulate Bacterial cGAS-like Enzymes to Mediate Bacteriophage Immunity.
Mol.Cell, 77, 2020
|
|
6DEI
 
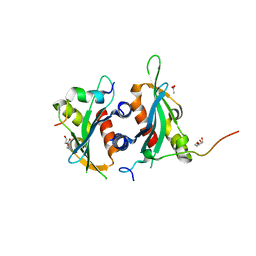 | | Structure of Dse3-Csm1 complex | | Descriptor: | ACETATE ION, Monopolin complex subunit CSM1, Protein DSE3, ... | | Authors: | Singh, N, Corbett, K.D. | | Deposit date: | 2018-05-12 | | Release date: | 2018-10-03 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.699 Å) | | Cite: | The budding-yeast RWD protein Csm1 scaffolds diverse protein complexes through a conserved structural mechanism.
Protein Sci., 27, 2018
|
|
6DD8
 
 | | Structure of mouse SYCP3, P21 form | | Descriptor: | Synaptonemal complex protein 3 | | Authors: | Rosenberg, S.C, Munoz, I.C, Uson, I, Corbett, K.D. | | Deposit date: | 2018-05-09 | | Release date: | 2018-08-01 | | Last modified: | 2020-01-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A conserved filamentous assembly underlies the structure of the meiotic chromosome axis.
Elife, 8, 2019
|
|
6DEX
 
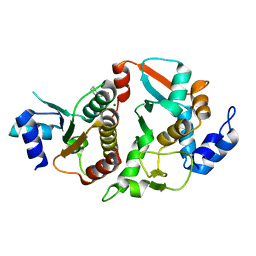 | | Structure of Eremothecium gossypii Shu1:Shu2 complex | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, MAGNESIUM ION, Suppressor of hydroxyurea sensitivity protein 1, ... | | Authors: | Singh, N, Corbett, K.D. | | Deposit date: | 2018-05-13 | | Release date: | 2018-05-23 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of Eremothecium gossypii Shu1:Shu2 complex
To Be Published
|
|
1TOK
 
 | | Maleic acid-bound structure of SRHEPT mutant of E. coli aspartate aminotransferase | | Descriptor: | Aspartate aminotransferase, MALEIC ACID | | Authors: | Chow, M.A, McElroy, K.E, Corbett, K.D, Berger, J.M, Kirsch, J.F. | | Deposit date: | 2004-06-14 | | Release date: | 2004-10-05 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Narrowing substrate specificity in a directly evolved enzyme: the A293D mutant of aspartate aminotransferase
Biochemistry, 43, 2004
|
|
1TOJ
 
 | | Hydrocinnamic acid-bound structure of SRHEPT mutant of E. coli aspartate aminotransferase | | Descriptor: | Aspartate aminotransferase, HYDROCINNAMIC ACID | | Authors: | Chow, M.A, McElroy, K.E, Corbett, K.D, Berger, J.M, Kirsch, J.F. | | Deposit date: | 2004-06-14 | | Release date: | 2004-10-05 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Narrowing substrate specificity in a directly evolved enzyme: the A293D mutant of aspartate aminotransferase
Biochemistry, 43, 2004
|
|
1TOE
 
 | | Unliganded structure of Hexamutant + A293D mutant of E. coli aspartate aminotransferase | | Descriptor: | Aspartate aminotransferase, SULFATE ION | | Authors: | Chow, M.A, McElroy, K.E, Corbett, K.D, Berger, J.M, Kirsch, J.F. | | Deposit date: | 2004-06-14 | | Release date: | 2004-10-05 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Narrowing substrate specificity in a directly evolved enzyme: the A293D mutant of aspartate aminotransferase
Biochemistry, 43, 2004
|
|
1TOG
 
 | | Hydrocinnamic acid-bound structure of SRHEPT + A293D mutant of E. coli aspartate aminotransferase | | Descriptor: | Aspartate aminotransferase, HYDROCINNAMIC ACID | | Authors: | Chow, M.A, McElroy, K.E, Corbett, K.D, Berger, J.M, Kirsch, J.F. | | Deposit date: | 2004-06-14 | | Release date: | 2004-10-05 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Narrowing substrate specificity in a directly evolved enzyme: the A293D mutant of aspartate aminotransferase
Biochemistry, 43, 2004
|
|
1TOI
 
 | | Hydrocinnamic acid-bound structure of Hexamutant + A293D mutant of E. coli aspartate aminotransferase | | Descriptor: | Aspartate aminotransferase, HYDROCINNAMIC ACID | | Authors: | Chow, M.A, McElroy, K.E, Corbett, K.D, Berger, J.M, Kirsch, J.F. | | Deposit date: | 2004-06-14 | | Release date: | 2004-10-05 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Narrowing substrate specificity in a directly evolved enzyme: the A293D mutant of aspartate aminotransferase
Biochemistry, 43, 2004
|
|
5VQ9
 
 | | Structure of human TRIP13, Apo form | | Descriptor: | Pachytene checkpoint protein 2 homolog | | Authors: | Ye, Q, Corbett, K.D. | | Deposit date: | 2017-05-08 | | Release date: | 2017-06-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.02 Å) | | Cite: | The AAA+ ATPase TRIP13 remodels HORMA domains through N-terminal engagement and unfolding.
EMBO J., 36, 2017
|
|
5V3N
 
 | | Structure of S. cerevisiae Ulp2-Tof2-Csm1 complex | | Descriptor: | Monopolin complex subunit CSM1, Ulp2p,Topoisomerase 1-associated factor 2 chimera | | Authors: | SIngh, N, Corbett, K.D. | | Deposit date: | 2017-03-07 | | Release date: | 2017-05-17 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Recruitment of a SUMO isopeptidase to rDNA stabilizes silencing complexes by opposing SUMO targeted ubiquitin ligase activity.
Genes Dev., 31, 2017
|
|
5V1A
 
 | | Structure of S. cerevisiae Ulp2:Csm1 complex | | Descriptor: | Monopolin complex subunit CSM1, Ubiquitin-like-specific protease 2 | | Authors: | Singh, N, Corbett, K.D. | | Deposit date: | 2017-03-01 | | Release date: | 2017-05-17 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Recruitment of a SUMO isopeptidase to rDNA stabilizes silencing complexes by opposing SUMO targeted ubiquitin ligase activity.
Genes Dev., 31, 2017
|
|
5VQA
 
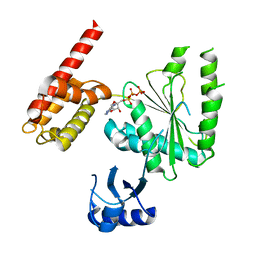 | | Structure of human TRIP13, ATP-bound form | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Pachytene checkpoint protein 2 homolog | | Authors: | Ye, Q, Corbett, K.D. | | Deposit date: | 2017-05-08 | | Release date: | 2017-06-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | The AAA+ ATPase TRIP13 remodels HORMA domains through N-terminal engagement and unfolding.
EMBO J., 36, 2017
|
|
6P7P
 
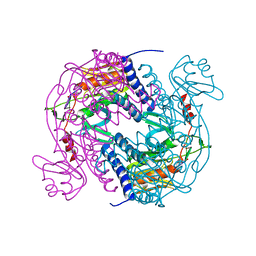 | | Structure of E. coli MS115-1 NucC, cAAA-bound form | | Descriptor: | CHLORIDE ION, Cyclic tri-AMP (5'-3' linked), E. coli MS115-1 NucC 2-241 | | Authors: | Ye, Q, Lau, R.K, Berg, K.R, Corbett, K.D. | | Deposit date: | 2019-06-06 | | Release date: | 2019-12-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.665 Å) | | Cite: | Structure and Mechanism of a Cyclic Trinucleotide-Activated Bacterial Endonuclease Mediating Bacteriophage Immunity.
Mol.Cell, 77, 2020
|
|
6P80
 
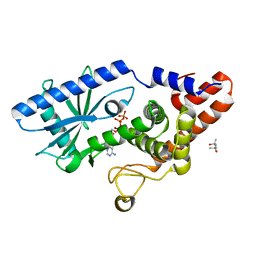 | | Structure of E. coli MS115-1 CdnC + ATP | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ADENOSINE-5'-TRIPHOSPHATE, CHLORIDE ION, ... | | Authors: | Ye, Q, Azimi, C.S, Corbett, K.D. | | Deposit date: | 2019-06-06 | | Release date: | 2019-12-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | HORMA Domain Proteins and a Trip13-like ATPase Regulate Bacterial cGAS-like Enzymes to Mediate Bacteriophage Immunity.
Mol.Cell, 77, 2020
|
|
6P8P
 
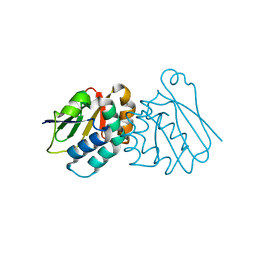 | | Structure of P. aeruginosa ATCC27853 HORMA1 | | Descriptor: | CALCIUM ION, Uncharacterized protein | | Authors: | Ye, Q, Corbett, K.D. | | Deposit date: | 2019-06-07 | | Release date: | 2019-12-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.635 Å) | | Cite: | HORMA Domain Proteins and a Trip13-like ATPase Regulate Bacterial cGAS-like Enzymes to Mediate Bacteriophage Immunity.
Mol.Cell, 77, 2020
|
|
6P8V
 
 | | Structure of E. coli MS115-1 HORMA:CdnC:Trip13 complex | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, ATPase, AAA family, ... | | Authors: | Ye, Q, Corbett, K.D. | | Deposit date: | 2019-06-08 | | Release date: | 2019-12-25 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | HORMA Domain Proteins and a Trip13-like ATPase Regulate Bacterial cGAS-like Enzymes to Mediate Bacteriophage Immunity.
Mol.Cell, 77, 2020
|
|
6P8R
 
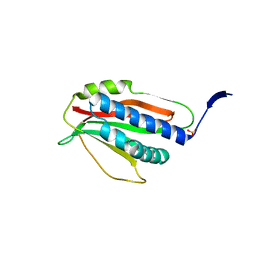 | | Structure of P. aeruginosa ATCC27853 HORMA2 | | Descriptor: | HORMA domain containing protein, PHOSPHATE ION | | Authors: | Ye, Q, Corbett, K.D. | | Deposit date: | 2019-06-07 | | Release date: | 2019-12-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.141 Å) | | Cite: | HORMA Domain Proteins and a Trip13-like ATPase Regulate Bacterial cGAS-like Enzymes to Mediate Bacteriophage Immunity.
Mol.Cell, 77, 2020
|
|
6PB3
 
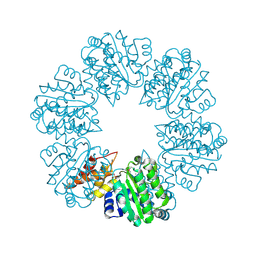 | | Structure of Rhizobiales Trip13 | | Descriptor: | Rhizobiales Sp. Pch2, SULFATE ION | | Authors: | Ye, Q, Corbett, K.D. | | Deposit date: | 2019-06-12 | | Release date: | 2019-12-25 | | Last modified: | 2020-03-04 | | Method: | X-RAY DIFFRACTION (2.048 Å) | | Cite: | HORMA Domain Proteins and a Trip13-like ATPase Regulate Bacterial cGAS-like Enzymes to Mediate Bacteriophage Immunity.
Mol.Cell, 77, 2020
|
|
6P82
 
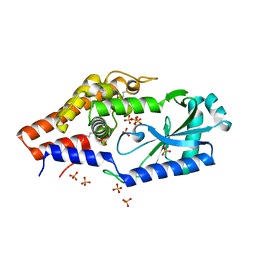 | | Structure of P. aeruginosa ATCC27853 CdnD, Apo form 1 | | Descriptor: | Nucleotidyltransferase, SULFATE ION | | Authors: | Ye, Q, Corbett, K.D. | | Deposit date: | 2019-06-06 | | Release date: | 2019-12-25 | | Last modified: | 2020-03-04 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | HORMA Domain Proteins and a Trip13-like ATPase Regulate Bacterial cGAS-like Enzymes to Mediate Bacteriophage Immunity.
Mol.Cell, 77, 2020
|
|
