8F44
 
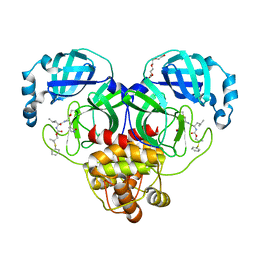 | | Crystal structure of SARS-CoV-2 3CL protease in complex with a dimethyl phenyl sulfane inhibitor | | 分子名称: | (1R,2S)-1-hydroxy-2-[(N-{[2-methyl-2-(phenylsulfanyl)propoxy]carbonyl}-L-leucyl)amino]-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, (2-methyl-2-phenylsulfanyl-propyl) ~{N}-[(2~{S})-1-[[(1~{S},2~{S})-1-[bis(oxidanyl)-oxidanylidene-$l^{5}-sulfanyl]-1-oxidanyl-3-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]amino]-4-methyl-1-oxidanylidene-pentan-2-yl]carbamate, 3C-like proteinase, ... | | 著者 | Liu, L, Lovell, S, Cooper, A, Battaile, K.P, Dampalla, C.S, Groutas, W.C. | | 登録日 | 2022-11-10 | | 公開日 | 2022-11-23 | | 最終更新日 | 2024-10-09 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Structure-guided design of direct-acting antivirals that exploit the gem-dimethyl effect and potently inhibit 3CL proteases of severe acute respiratory syndrome Coronavirus-2 (SARS-CoV-2) and middle east respiratory syndrome coronavirus (MERS-CoV).
Eur.J.Med.Chem., 254, 2023
|
|
5HD7
 
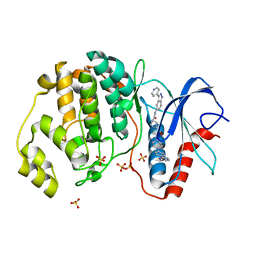 | | Dissecting Therapeutic Resistance to ERK Inhibition Rat Mutant SCH772984 in complex with (3R)-1-(2-oxo-2-{4-[4-(pyrimidin-2-yl)phenyl]piperazin-1-yl}ethyl)-N-[3-(pyridin-4-yl)-2H-indazol-5-yl]pyrrolidine-3-carboxamide | | 分子名称: | (3R)-1-(2-oxo-2-{4-[4-(pyrimidin-2-yl)phenyl]piperazin-1-yl}ethyl)-N-[3-(pyridin-4-yl)-2H-indazol-5-yl]pyrrolidine-3-carboxamide, Mitogen-activated protein kinase 1, SULFATE ION | | 著者 | Jha, S, Morris, E.J, Hruza, A, Mansueto, M.S, Schroeder, G, Arbanas, J, McMasters, D, Restaino, C.R, Dayananth, R, Black, S, Elsen, N.L, Mannarino, A, Cooper, A, Fawell, S, Zawel, L, Jayaraman, L, Samatar, A.A. | | 登録日 | 2016-01-04 | | 公開日 | 2016-02-24 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (1.69 Å) | | 主引用文献 | Dissecting Therapeutic Resistance to ERK Inhibition.
Mol.Cancer Ther., 15, 2016
|
|
5HD4
 
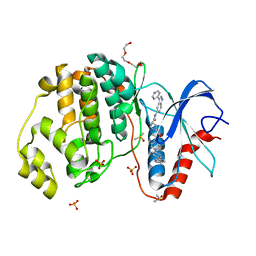 | | Dissecting Therapeutic Resistance to ERK Inhibition Rat Wild Type SCH772984 in complex with (3R)-1-(2-oxo-2-{4-[4-(pyrimidin-2-yl)phenyl]piperazin-1-yl}ethyl)-N-[3-(pyridin-4-yl)-2H-indazol-5-yl]pyrrolidine-3-carboxamide | | 分子名称: | (3R)-1-(2-oxo-2-{4-[4-(pyrimidin-2-yl)phenyl]piperazin-1-yl}ethyl)-N-[3-(pyridin-4-yl)-2H-indazol-5-yl]pyrrolidine-3-carboxamide, DIMETHYL SULFOXIDE, Mitogen-activated protein kinase 1, ... | | 著者 | Jha, S, Morris, E.J, Hruza, A, Mansueto, M.S, Schroeder, G, Arbanas, J, McMasters, D, Restaino, C.R, Dayananth, R, Black, S, Elsen, N.L, Mannarino, A, Cooper, A, Fawell, S, Zawel, L, Jayaraman, L, Samatar, A.A. | | 登録日 | 2016-01-04 | | 公開日 | 2016-02-24 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.45 Å) | | 主引用文献 | Dissecting Therapeutic Resistance to ERK Inhibition.
Mol.Cancer Ther., 15, 2016
|
|
1XGK
 
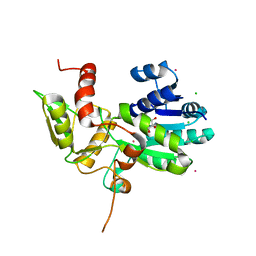 | | CRYSTAL STRUCTURE OF N12G AND A18G MUTANT NMRA | | 分子名称: | CHLORIDE ION, GLYCEROL, NITROGEN METABOLITE REPRESSION REGULATOR NMRA, ... | | 著者 | Lamb, H.K, Ren, J, Park, A, Johnson, C, Leslie, K, Cocklin, S, Thompson, P, Mee, C, Cooper, A, Stammers, D.K, Hawkins, A.R. | | 登録日 | 2004-09-17 | | 公開日 | 2004-12-14 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Modulation of the ligand binding properties of the transcription repressor NmrA by GATA-containing DNA and site-directed mutagenesis
Protein Sci., 13, 2004
|
|
5AE8
 
 | | Crystal structure of mouse PI3 kinase delta in complex with GSK2269557 | | 分子名称: | 6-(1H-Indol-4-yl)-4-(5-{[4-(1-methylethyl)-1-piperazinyl]methyl}-1,3-oxazol-2-yl)-1H-indazole, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit delta isoform | | 著者 | Down, K.D, Amour, A, Baldwin, I.R, Cooper, A.W.J, Deakin, A.M, Felton, L.M, Guntrip, S.B, Hardy, C, Harrison, Z.A, Jones, K.L, Jones, P, Keeling, S.E, Le, J, Livia, S, Lucas, F, Lunniss, C.J, Parr, N.J, Robinson, E, Rowland, P, Smith, S, Thomas, D.A, Vitulli, G, Washio, Y, Hamblin, N. | | 登録日 | 2015-08-26 | | 公開日 | 2015-09-16 | | 最終更新日 | 2024-06-19 | | 実験手法 | X-RAY DIFFRACTION (2.42 Å) | | 主引用文献 | Optimization of Novel Indazoles as Highly Potent and Selective Inhibitors of Phosphoinositide 3-Kinase Gamma for the Treatment of Respiratory Disease.
J.Med.Chem., 58, 2015
|
|
5AE9
 
 | | Crystal structure of mouse PI3 kinase delta in complex with GSK2292767 | | 分子名称: | N-[5-[4-(5-{[(2R,6S)-2,6-DIMETHYL-4-MORPHOLINYL]METHYL}-1,3-OXAZOL-2-YL)-1H-INDAZOL-6-YL]-2-(METHYLOXY)-3-PYRIDINYL]METHANESULFONAMIDE, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit delta isoform | | 著者 | Down, K.D, Amour, A, Baldwin, I.R, Cooper, A.W.J, Deakin, A.M, Felton, L.M, Guntrip, S.B, Hardy, C, Harrison, Z.A, Jones, K.L, Jones, P, Keeling, S.E, Le, J, Livia, S, Lucas, F, Lunniss, C.J, Parr, N.J, Robinson, E, Rowland, P, Smith, S, Thomas, D.A, Vitulli, G, Washio, Y, Hamblin, N. | | 登録日 | 2015-08-26 | | 公開日 | 2015-09-16 | | 最終更新日 | 2024-06-19 | | 実験手法 | X-RAY DIFFRACTION (2.44 Å) | | 主引用文献 | Optimization of Novel Indazoles as Highly Potent and Selective Inhibitors of Phosphoinositide 3-Kinase Gamma for the Treatment of Respiratory Disease.
J.Med.Chem., 58, 2015
|
|
3ZPM
 
 | | Solution structure of latherin | | 分子名称: | LATHERIN | | 著者 | Vance, S.J, MacDonald, R.E, Cooper, A, Kennedy, M.W, Smith, B.O. | | 登録日 | 2013-02-28 | | 公開日 | 2013-06-26 | | 最終更新日 | 2023-06-14 | | 実験手法 | SOLUTION NMR | | 主引用文献 | The structure of latherin, a surfactant allergen protein from horse sweat and saliva.
J R Soc Interface, 10, 2013
|
|
2XV9
 
 | | The solution structure of ABA-1A saturated with oleic acid | | 分子名称: | ABA-1A1 REPEAT UNIT | | 著者 | Smith, B.O, Kennedy, M.W, Cooper, A, Meenan, N.A.G, Bromek, K. | | 登録日 | 2010-10-25 | | 公開日 | 2011-08-10 | | 最終更新日 | 2023-06-14 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of a repeated unit of the ABA-1 nematode polyprotein allergen of Ascaris reveals a novel fold and two discrete lipid-binding sites.
PLoS Negl Trop Dis, 5, 2011
|
|
1XBD
 
 | | INTERNAL XYLAN BINDING DOMAIN FROM CELLULOMONAS FIMI XYLANASE D, NMR, 5 STRUCTURES | | 分子名称: | XYLANASE D | | 著者 | Simpson, P.J, Bolam, D.N, Cooper, A, Ciruela, A, Hazlewood, G.P, Gilbert, H.J, Williamson, M.P. | | 登録日 | 1998-10-16 | | 公開日 | 1999-07-21 | | 最終更新日 | 2022-03-02 | | 実験手法 | SOLUTION NMR | | 主引用文献 | A family IIb xylan-binding domain has a similar secondary structure to a homologous family IIa cellulose-binding domain but different ligand specificity.
Structure Fold.Des., 7, 1999
|
|
3ZS2
 
 | | TyrB25,NMePheB26,LysB28,ProB29-insulin analogue crystal structure | | 分子名称: | CHLORIDE ION, INSULIN A CHAIN, INSULIN B CHAIN, ... | | 著者 | Antolikova, E, Zakova, L, Turkenburg, J.P, Watson, C.J, Hanclova, I, Sanda, M, Cooper, A, Kraus, T, Brzozowski, A.M, Jiracek, J.A. | | 登録日 | 2011-06-21 | | 公開日 | 2011-08-31 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.97 Å) | | 主引用文献 | Non-Equivalent Role of Inter- and Intramolecular Hydrogen Bonds in the Insulin Dimer Interface.
J.Biol.Chem., 286, 2011
|
|
2VH3
 
 | | ranasmurfin | | 分子名称: | GLYCEROL, RANASMURFIN, SULFATE ION, ... | | 著者 | Oke, M, Ching, R.T, Carter, L.G, Johnson, K.A, Liu, H, McMahon, S.A, Bloch Junior, C, Botting, C.H, Walsh, M.A, Latiff, A.A, Kennedy, M.W, Cooper, A, Naismith, J.H. | | 登録日 | 2007-11-17 | | 公開日 | 2007-12-04 | | 最終更新日 | 2011-08-31 | | 実験手法 | X-RAY DIFFRACTION (1.16 Å) | | 主引用文献 | Unusual Chromophore and Cross-Links in Ranasmurfin: A Blue Protein from the Foam Nests of a Tropical Frog.
Angew.Chem.Int.Ed.Engl., 47, 2008
|
|
1TI7
 
 | | CRYSTAL STRUCTURE OF NMRA, A NEGATIVE TRANSCRIPTIONAL REGULATOR, IN COMPLEX WITH NADP AT 1.7A RESOLUTION | | 分子名称: | CHLORIDE ION, GLYCEROL, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | 著者 | Lamb, H.K, Leslie, K, Dodds, A.L, Nutley, M, Cooper, A, Johnson, C, Thompson, P, Stammers, D.K, Hawkins, A.R. | | 登録日 | 2004-06-02 | | 公開日 | 2004-06-08 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | The negative transcriptional regulator NmrA discriminates between oxidized and reduced dinucleotides.
J.Biol.Chem., 278, 2003
|
|
9DN4
 
 | | Crystal structure of a SARS-CoV-2 20-mer RNA in complex with FAB BL3-6S97N | | 分子名称: | CHLORIDE ION, FAB BL3-6S97N HEAVY CHAIN, FAB BL3-6S97N LIGHT CHAIN, ... | | 著者 | Lovell, S, Cooper, A, Battaile, K.P, Hegde, S, Wang, J. | | 登録日 | 2024-09-16 | | 公開日 | 2024-09-25 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Crystal structure of a SARS-CoV-2 20-mer RNA in complex with FAB BL3-6S97N
To be published
|
|
2XBD
 
 | | INTERNAL XYLAN BINDING DOMAIN FROM CELLULOMONAS FIMI XYLANASE D, NMR, MINIMIZED AVERAGE STRUCTURE | | 分子名称: | XYLANASE D | | 著者 | Simpson, P.J, Bolam, D.N, Cooper, A, Ciruela, A, Hazlewood, G.P, Gilbert, H.J, Williamson, M.P. | | 登録日 | 1998-10-27 | | 公開日 | 1999-07-21 | | 最終更新日 | 2022-03-16 | | 実験手法 | SOLUTION NMR | | 主引用文献 | A family IIb xylan-binding domain has a similar secondary structure to a homologous family IIa cellulose-binding domain but different ligand specificity.
Structure Fold.Des., 7, 1999
|
|
2WGO
 
 | | Structure of ranaspumin-2, a surfactant protein from the foam nests of a tropical frog | | 分子名称: | RANASPUMIN-2 | | 著者 | Mackenzie, C.D, Smith, B.O, Kennedy, M.W, Cooper, A. | | 登録日 | 2009-04-21 | | 公開日 | 2009-06-23 | | 最終更新日 | 2011-07-13 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Ranaspumin-2: Structure and Function of a Surfactant Protein from the Foam Nests of a Tropical Frog.
Biophys.J., 96, 2009
|
|
6NTY
 
 | | 2.1 A resolution structure of the Musashi-2 (Msi2) RNA recognition motif 1 (RRM1) domain | | 分子名称: | PHOSPHATE ION, RNA-binding protein Musashi homolog 2 | | 著者 | Lovell, S, Kashipathy, M.M, Battaile, K.P, Lan, L, Xiaoqing, W, Cooper, A, Gao, F.P, Xu, L. | | 登録日 | 2019-01-30 | | 公開日 | 2019-10-23 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Crystal and solution structures of human oncoprotein Musashi-2 N-terminal RNA recognition motif 1.
Proteins, 88, 2020
|
|
3ZQR
 
 | | NMePheB25 insulin analogue crystal structure | | 分子名称: | CHLORIDE ION, INSULIN A CHAIN, INSULIN B CHAIN, ... | | 著者 | Antolikova, E, Zakova, L, Turkenburg, J.P, Watson, C.J, Hanclova, I, Sanda, M, Cooper, A, Kraus, T, Brzozowski, A.M, Jiracek, J.A. | | 登録日 | 2011-06-10 | | 公開日 | 2011-08-24 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Non-Equivalent Role of Inter- and Intramolecular Hydrogen Bonds in the Insulin Dimer Interface.
J.Biol.Chem., 286, 2011
|
|
5HKW
 
 | | Crystal Structure of Apo c-Cbl TKBD Refined to 2.25 A Resolution | | 分子名称: | E3 ubiquitin-protein ligase CBL, SODIUM ION | | 著者 | Lovell, S, Battaile, K.P, Mehzabeen, N, Zhang, N, Cooper, A, Gao, P, Perez, R.P. | | 登録日 | 2016-01-14 | | 公開日 | 2017-01-18 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | Crystal Structure of Apo c-Cbl TKBD Refined to 2.25 A Resolution
To be published
|
|
5HKZ
 
 | | Crystal Structure of c-Cbl TKBD in complex with SPRY2 peptide (36-60, pY55) Refined to 1.8 A Resolution (P21 form) | | 分子名称: | E3 ubiquitin-protein ligase CBL, Protein sprouty homolog 2, SODIUM ION | | 著者 | Lovell, S, Battaile, K.P, Mehzabeen, N, Zhang, N, Cooper, A, Gao, P, Perez, R.P. | | 登録日 | 2016-01-14 | | 公開日 | 2017-01-18 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Crystal Structure of c-Cbl TKBD in complex with SPRY2 peptide (36-60, pY55) Refined to 1.8 A Resolution (P21 form)
To be published
|
|
5HKX
 
 | | Crystal Structure of c-Cbl TKBD-RING domains (Y371E mutant) Refined to 1.85 A Resolution | | 分子名称: | 1,2-ETHANEDIOL, E3 ubiquitin-protein ligase CBL, SODIUM ION, ... | | 著者 | Lovell, S, Battaile, K.P, Mehzabeen, N, Zhang, N, Cooper, A, Gao, P, Perez, R.P. | | 登録日 | 2016-01-14 | | 公開日 | 2017-01-18 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Crystal Structure of c-Cbl TKBD-RING domains (Y371E mutant) Refined to 1.85 A Resolution
To be published
|
|
5HKY
 
 | | Crystal structure of c-Cbl TKBD domain in complex with SPRY2 peptide (36-60, pY55) Refined to 1.8A Resolution (P6 form) | | 分子名称: | CHLORIDE ION, E3 ubiquitin-protein ligase CBL, PENTAETHYLENE GLYCOL, ... | | 著者 | Lovell, S, Battaile, K.P, Mehzabeen, N, Zhang, N, Cooper, A, Gao, P, Perez, R.P. | | 登録日 | 2016-01-14 | | 公開日 | 2017-01-18 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Crystal structure of c-Cbl TKBD domain in complex with SPRY2 peptide (36-60, pY55) Refined to 1.8A Resolution (P6 form)
To be published
|
|
5HL0
 
 | | Crystal Structure of c-Cbl TKBD in complex with SPRY2 peptide (54-60, pY55) Refined to 2.2A Resolution | | 分子名称: | E3 ubiquitin-protein ligase CBL, SODIUM ION, Sprouty 2 (SPRY2) | | 著者 | Lovell, S, Battaile, K.P, Mehzabeen, N, Zhang, N, Cooper, A, Gao, P, Perez, R.P. | | 登録日 | 2016-01-14 | | 公開日 | 2017-01-18 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Crystal Structure of c-Cbl TKBD in complex with SPRY2 peptide (54-60, pY55) Refined to 2.2A Resolution
To Be Published
|
|
