1EQB
 
 | | X-RAY CRYSTAL STRUCTURE AT 2.7 ANGSTROMS RESOLUTION OF TERNARY COMPLEX BETWEEN THE Y65F MUTANT OF E-COLI SERINE HYDROXYMETHYLTRANSFERASE, GLYCINE AND 5-FORMYL TETRAHYDROFOLATE | | 分子名称: | GLYCINE, N-[4-({[(6S)-2-amino-5-formyl-4-oxo-3,4,5,6,7,8-hexahydropteridin-6-yl]methyl}amino)benzoyl]-L-glutamic acid, PYRIDOXAL-5'-PHOSPHATE, ... | | 著者 | Contestabile, R, Angelaccio, S, Bossa, F, Wright, H.T, Scarsdale, N, Kazanina, G, Schirch, V. | | 登録日 | 2000-04-03 | | 公開日 | 2000-04-19 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Role of tyrosine 65 in the mechanism of serine hydroxymethyltransferase.
Biochemistry, 39, 2000
|
|
2MMZ
 
 | | Solution structure of the apo form of human glutaredoxin 5 | | 分子名称: | Glutaredoxin-related protein 5, mitochondrial | | 著者 | Banci, L, Brancaccio, D, Ciofi-Baffoni, S, Del Conte, R, Gadepalli, R, Mikolajczyk, M, Neri, S, Piccioli, M, Winkelmann, J. | | 登録日 | 2014-03-25 | | 公開日 | 2014-04-16 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | [2Fe-2S] cluster transfer in iron-sulfur protein biogenesis.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
1L3N
 
 | | The Solution Structure of Reduced Dimeric Copper Zinc SOD: the Structural Effects of Dimerization | | 分子名称: | COPPER (I) ION, ZINC ION, superoxide dismutase [Cu-Zn] | | 著者 | Banci, L, Bertini, I, Cramaro, F, Del Conte, R, Viezzoli, M.S. | | 登録日 | 2002-02-28 | | 公開日 | 2002-05-08 | | 最終更新日 | 2021-10-27 | | 実験手法 | SOLUTION NMR | | 主引用文献 | The solution structure of reduced dimeric copper zinc superoxide dismutase. The structural effects of dimerization
Eur.J.Biochem., 269, 2002
|
|
1HKF
 
 | | The three dimensional structure of NK cell receptor Nkp44, a triggering partner in natural cytotoxicity | | 分子名称: | NK CELL ACTIVATING RECEPTOR | | 著者 | Ponassi, M, Cantoni, C, Biassoni, R, Conte, R, Spallarossa, A, Moretta, A, Moretta, L, Bolognesi, M, Bordo, D. | | 登録日 | 2003-03-10 | | 公開日 | 2003-06-11 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | The Three-Dimensional Structure of the Human Nk Cell Receptor Nkp44, a Triggering Partner in Natural Cytotoxicity
Structure, 11, 2003
|
|
1K0V
 
 | | Copper trafficking: the solution structure of Bacillus subtilis CopZ | | 分子名称: | COPPER (I) ION, CopZ | | 著者 | Banci, L, Bertini, I, Del Conte, R, Markey, J, Ruiz-Duenas, F.J. | | 登録日 | 2001-09-21 | | 公開日 | 2001-12-19 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Copper trafficking: the solution structure of Bacillus subtilis CopZ.
Biochemistry, 40, 2001
|
|
1OLL
 
 | | Extracellular region of the human receptor NKp46 | | 分子名称: | 1,2-ETHANEDIOL, NK RECEPTOR | | 著者 | Ponassi, M, Cantoni, C, Biassoni, R, Conte, R, Spallarossa, A, Pesce, A, Moretta, A, Moretta, L, Bolognesi, M, Bordo, D. | | 登録日 | 2003-08-07 | | 公開日 | 2003-09-04 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (1.93 Å) | | 主引用文献 | Structure of the Human Nk Cell Triggering Receptor Nkp46 Ectodomain
Biochem.Biophys.Res.Commun., 309, 2003
|
|
1DSW
 
 | | THE SOLUTION STRUCTURE OF A MONOMERIC, REDUCED FORM OF HUMAN COPPER, ZINC SUPEROXIDE DISMUTASE BEARING THE SAME CHARGE AS THE NATIVE PROTEIN | | 分子名称: | COPPER (II) ION, SUPEROXIDE DISMUTASE (CU-ZN), ZINC ION | | 著者 | Banci, L, Bertini, I, Del Conte, R, Fadin, R, Mangani, S, Viezzoli, M.S. | | 登録日 | 2000-01-10 | | 公開日 | 2000-03-22 | | 最終更新日 | 2022-02-16 | | 実験手法 | SOLUTION NMR | | 主引用文献 | The solution structure of a monomeric, reduced form of human copper,zinc superoxide dismutase bearing the same charge as the native protein.
J.Biol.Inorg.Chem., 4, 1999
|
|
2N3Y
 
 | | NMR structure of the Y48pCMF variant of human cytochrome c in its reduced state | | 分子名称: | Cytochrome c, Mesoheme | | 著者 | Moreno-Beltran, B, Del Conte, R, Diaz-Quintana, A, De la Rosa, M.A, Turano, P, Diaz-Moreno, I. | | 登録日 | 2015-06-15 | | 公開日 | 2016-12-14 | | 最終更新日 | 2017-04-26 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structural basis of mitochondrial dysfunction in response to cytochrome c phosphorylation at tyrosine 48.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
1BA9
 
 | | THE SOLUTION STRUCTURE OF REDUCED MONOMERIC SUPEROXIDE DISMUTASE, NMR, 36 STRUCTURES | | 分子名称: | COPPER (I) ION, SUPEROXIDE DISMUTASE, ZINC ION | | 著者 | Banci, L, Benedetto, M, Bertini, I, Del Conte, R, Piccioli, M, Viezzoli, M.S. | | 登録日 | 1998-04-24 | | 公開日 | 1998-09-16 | | 最終更新日 | 2021-11-03 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of reduced monomeric Q133M2 copper, zinc superoxide dismutase (SOD). Why is SOD a dimeric enzyme?.
Biochemistry, 37, 1998
|
|
2ORL
 
 | | Solution structure of the cytochrome c- para-aminophenol adduct | | 分子名称: | 4-AMINOPHENOL, Cytochrome c iso-1, HEME C | | 著者 | Assfalg, M, Bertini, I, Del Conte, R, Giachetti, A, Turano, P. | | 登録日 | 2007-02-03 | | 公開日 | 2007-04-24 | | 最終更新日 | 2021-10-20 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Cytochrome c and organic molecules: solution structure of the p-aminophenol adduct.
Biochemistry, 46, 2007
|
|
1S4I
 
 | | Crystal structure of a SOD-like protein from Bacillus subtilis | | 分子名称: | CHLORIDE ION, ZINC ION, superoxide dismutase-like protein yojM | | 著者 | Banci, L, Bertini, I, Calderone, V, Cramaro, F, Del Conte, R, Fantoni, A, Mangani, S, Quattrone, A, Viezzoli, M.S. | | 登録日 | 2004-01-16 | | 公開日 | 2005-04-26 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | A prokaryotic superoxide dismutase paralog lacking two Cu ligands: from largely unstructured in solution to ordered in the crystal.
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
1P8G
 
 | |
2JSD
 
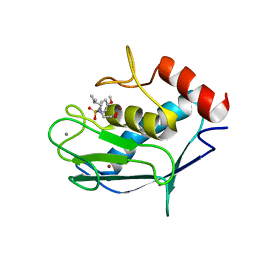 | | Solution structure of MMP20 complexed with NNGH | | 分子名称: | CALCIUM ION, Matrix metalloproteinase-20, N-ISOBUTYL-N-[4-METHOXYPHENYLSULFONYL]GLYCYL HYDROXAMIC ACID, ... | | 著者 | Arendt, Y, Banci, L, Bertini, I, Cantini, F, Cozzi, R, Del Conte, R, Gonnelli, L, Structural Proteomics in Europe (SPINE) | | 登録日 | 2007-07-03 | | 公開日 | 2007-11-20 | | 最終更新日 | 2024-05-29 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Catalytic domain of MMP20 (Enamelysin) - the NMR structure of a new matrix metalloproteinase.
Febs Lett., 581, 2007
|
|
2L1U
 
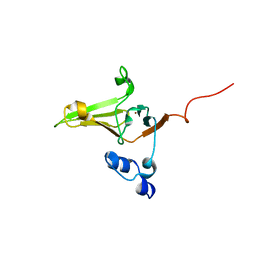 | | Structure-Functional Analysis of Mammalian MsrB2 protein | | 分子名称: | Methionine-R-sulfoxide reductase B2, mitochondrial, ZINC ION | | 著者 | Aachmann, F.L, Del Conte, R, Kwak, G, Kim, H, Gladyshev, V.N, Dikiy, A. | | 登録日 | 2010-08-06 | | 公開日 | 2010-08-18 | | 最終更新日 | 2024-05-01 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structure-Functional Analysis of Mammalian MsrB2 protein
To be Published
|
|
2HV4
 
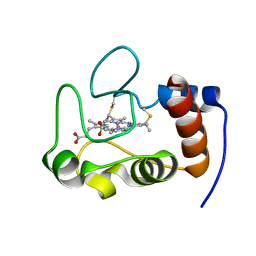 | | NMR solution structure refinement of yeast iso-1-ferrocytochrome c | | 分子名称: | Cytochrome c iso-1, HEME C | | 著者 | Assfalg, M, Bertini, I, Del Conte, R, Turano, P. | | 登録日 | 2006-07-27 | | 公開日 | 2006-09-26 | | 最終更新日 | 2021-10-20 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Cytochrome c and organic molecules: solution structure of the p-aminophenol adduct.
Biochemistry, 46, 2007
|
|
1U3N
 
 | | A SOD-like protein from B. subtilis, unstructured in solution, becomes ordered in the crystal: implications for function and for fibrillogenesis | | 分子名称: | Hypothetical superoxide dismutase-like protein yojM | | 著者 | Banci, L, Bertini, I, Calderone, V, Cramaro, F, Del Conte, R, Fantoni, A, Mangani, S, Quattrone, A, Viezzoli, M.S. | | 登録日 | 2004-07-22 | | 公開日 | 2005-05-03 | | 最終更新日 | 2022-03-02 | | 実験手法 | SOLUTION NMR | | 主引用文献 | A prokaryotic superoxide dismutase paralog lacking two Cu ligands: from largely unstructured in solution to ordered in the crystal.
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
1S6O
 
 | | Solution structure and backbone dynamics of the apo-form of the second metal-binding domain of the Menkes protein ATP7A | | 分子名称: | Copper-transporting ATPase 1 | | 著者 | Banci, L, Bertini, I, Del Conte, R, D'Onofrio, M, Rosato, A, Structural Proteomics in Europe (SPINE) | | 登録日 | 2004-01-26 | | 公開日 | 2004-04-06 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution Structure and Backbone Dynamics of the Cu(I) and Apo Forms of the Second Metal-Binding Domain of the Menkes Protein ATP7A.
Biochemistry, 43, 2004
|
|
1S6U
 
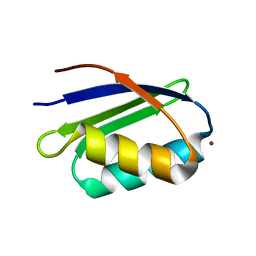 | | Solution structure and backbone dynamics of the Cu(I) form of the second metal-binding domain of the Menkes protein ATP7A | | 分子名称: | COPPER (I) ION, Copper-transporting ATPase 1 | | 著者 | Banci, L, Bertini, I, Del Conte, R, D'Onofrio, M, Rosato, A, Structural Proteomics in Europe (SPINE) | | 登録日 | 2004-01-27 | | 公開日 | 2004-04-06 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution Structure and Backbone Dynamics of the Cu(I) and Apo Forms of the Second Metal-Binding Domain of the Menkes Protein ATP7A.
Biochemistry, 43, 2004
|
|
1RK7
 
 | | Solution structure of apo Cu,Zn Superoxide Dismutase: role of metal ions in protein folding | | 分子名称: | Superoxide dismutase [Cu-Zn] | | 著者 | Banci, L, Bertini, I, Cramaro, F, Del Conte, R, Viezzoli, M.S. | | 登録日 | 2003-11-21 | | 公開日 | 2003-12-02 | | 最終更新日 | 2021-10-27 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of Apo Cu,Zn Superoxide Dismutase: Role of Metal Ions in Protein Folding
Biochemistry, 42, 2003
|
|
6YMF
 
 | | Crystal structure of serine hydroxymethyltransferase from Aphanothece halophytica in the PLP-Serine external aldimine state | | 分子名称: | GLYCEROL, PENTAETHYLENE GLYCOL, Serine hydroxymethyltransferase, ... | | 著者 | Ruszkowski, M, Sekula, B, Nogues, I, Tramonti, A, Angelaccio, S, Contestabile, R. | | 登録日 | 2020-04-08 | | 公開日 | 2020-06-03 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.63 Å) | | 主引用文献 | Structural and kinetic properties of serine hydroxymethyltransferase from the halophytic cyanobacterium Aphanothece halophytica provide a rationale for salt tolerance.
Int.J.Biol.Macromol., 159, 2020
|
|
6YME
 
 | | Crystal structure of serine hydroxymethyltransferase from Aphanothece halophytica in the PLP-internal aldimine state | | 分子名称: | DI(HYDROXYETHYL)ETHER, GLYCEROL, Serine hydroxymethyltransferase | | 著者 | Ruszkowski, M, Sekula, B, Nogues, I, Tramonti, A, Angelaccio, S, Contestabile, R. | | 登録日 | 2020-04-08 | | 公開日 | 2020-06-03 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.77 Å) | | 主引用文献 | Structural and kinetic properties of serine hydroxymethyltransferase from the halophytic cyanobacterium Aphanothece halophytica provide a rationale for salt tolerance.
Int.J.Biol.Macromol., 159, 2020
|
|
4PVF
 
 | | Crystal structure of Homo sapiens holo serine hydroxymethyltransferase 2 (mitochondrial) (SHMT2), isoform 3, transcript variant 5, 483 aa, at 2.6 ang. resolution | | 分子名称: | DI(HYDROXYETHYL)ETHER, GLYCEROL, Serine hydroxymethyltransferase, ... | | 著者 | Giardina, G, Brunotti, P, Fiascarelli, A, Contestabile, R, Cutruzzola, F. | | 登録日 | 2014-03-17 | | 公開日 | 2015-01-28 | | 最終更新日 | 2023-12-06 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | How pyridoxal 5'-phosphate differentially regulates human cytosolic and mitochondrial serine hydroxymethyltransferase oligomeric state.
Febs J., 282, 2015
|
|
6YMD
 
 | | Crystal structure of serine hydroxymethyltransferase from Aphanothece halophytica in the covalent complex with malonate | | 分子名称: | 1,2-ETHANEDIOL, 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, MALONATE ION, ... | | 著者 | Ruszkowski, M, Sekula, B, Nogues, I, Tramonti, A, Angelaccio, S, Contestabile, R. | | 登録日 | 2020-04-08 | | 公開日 | 2020-06-03 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.25 Å) | | 主引用文献 | Structural and kinetic properties of serine hydroxymethyltransferase from the halophytic cyanobacterium Aphanothece halophytica provide a rationale for salt tolerance.
Int.J.Biol.Macromol., 159, 2020
|
|
4LNM
 
 | | Structure of Escherichia coli Threonine Aldolase in Complex with Serine | | 分子名称: | (5-HYDROXY-4,6-DIMETHYLPYRIDIN-3-YL)METHYL DIHYDROGEN PHOSPHATE, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CALCIUM ION, ... | | 著者 | Safo, M.K, Contestabile, R, Remesh, S.G. | | 登録日 | 2013-07-11 | | 公開日 | 2013-11-06 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | On the catalytic mechanism and stereospecificity of Escherichia coli l-threonine aldolase.
Febs J., 281, 2014
|
|
4LNJ
 
 | | Structure of Escherichia coli Threonine Aldolase in Unliganded Form | | 分子名称: | (5-HYDROXY-4,6-DIMETHYLPYRIDIN-3-YL)METHYL DIHYDROGEN PHOSPHATE, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Low-specificity L-threonine aldolase, ... | | 著者 | Safo, M.K, Contestabile, R, Remesh, S.G. | | 登録日 | 2013-07-11 | | 公開日 | 2013-11-06 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | On the catalytic mechanism and stereospecificity of Escherichia coli l-threonine aldolase.
Febs J., 281, 2014
|
|
