5WDP
 
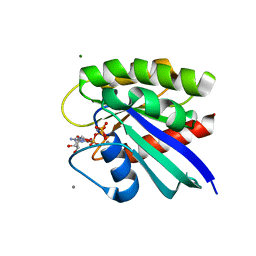 | | H-Ras mutant L120A bound to GMP-PNP at 277K | | Descriptor: | CALCIUM ION, GTPase HRas, MAGNESIUM ION, ... | | Authors: | Cofsky, J.C, Bandaru, P, Gee, C.L, Kuriyan, J. | | Deposit date: | 2017-07-05 | | Release date: | 2017-07-19 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Deconstruction of the Ras switching cycle through saturation mutagenesis.
Elife, 6, 2017
|
|
7S38
 
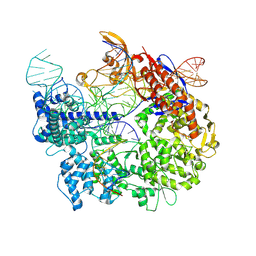 | | Cas9:sgRNA:DNA (S. pyogenes) forming a 3-base-pair R-loop | | Descriptor: | CRISPR-associated endonuclease Cas9/Csn1, Non-target DNA strand, Single-guide RNA, ... | | Authors: | Cofsky, J.C, Soczek, K.M, Knott, G.J, Nogales, E, Doudna, J.A. | | Deposit date: | 2021-09-04 | | Release date: | 2022-04-20 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | CRISPR-Cas9 bends and twists DNA to read its sequence.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7S37
 
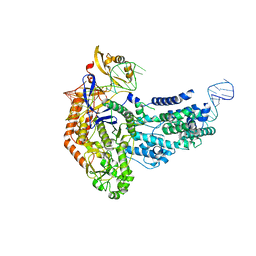 | | Cas9:sgRNA (S. pyogenes) in the open-protein conformation | | Descriptor: | CRISPR-associated endonuclease Cas9/Csn1, Single-guide RNA | | Authors: | Cofsky, J.C, Soczek, K.M, Knott, G.J, Nogales, E, Doudna, J.A. | | Deposit date: | 2021-09-04 | | Release date: | 2022-04-20 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | CRISPR-Cas9 bends and twists DNA to read its sequence.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7S3H
 
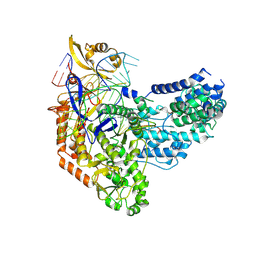 | | Cas9:sgRNA:DNA (S. pyogenes) with 0 RNA:DNA base pairs, open-protein/linear-DNA conformation | | Descriptor: | CRISPR-associated endonuclease Cas9/Csn1, Non-target DNA strand, Single-guide RNA, ... | | Authors: | Cofsky, J.C, Soczek, K.M, Knott, G.J, Nogales, E, Doudna, J.A. | | Deposit date: | 2021-09-06 | | Release date: | 2022-04-20 | | Last modified: | 2022-05-04 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | CRISPR-Cas9 bends and twists DNA to read its sequence.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7S36
 
 | | Cas9:sgRNA:DNA (S. pyogenes) with 0 RNA:DNA base pairs, closed-protein/bent-DNA conformation | | Descriptor: | CRISPR-associated endonuclease Cas9/Csn1, Non-target DNA strand, Single-guide RNA, ... | | Authors: | Cofsky, J.C, Soczek, K.M, Knott, G.J, Nogales, E, Doudna, J.A. | | Deposit date: | 2021-09-04 | | Release date: | 2022-04-20 | | Last modified: | 2022-05-04 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | CRISPR-Cas9 bends and twists DNA to read its sequence.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7THB
 
 | | Crystal structure of an RNA-5'/DNA-3' strand exchange junction | | Descriptor: | DNA (5'-D(*GP*AP*TP*GP*CP*TP*C)-3'), DNA (5'-D(*GP*TP*AP*AP*GP*CP*AP*GP*CP*AP*TP*C)-3'), RNA (5'-R(*AP*GP*CP*UP*UP*AP*C)-3') | | Authors: | Cofsky, J.C, Knott, G.J, Gee, C.L, Doudna, J.A. | | Deposit date: | 2022-01-10 | | Release date: | 2022-04-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Crystal structure of an RNA/DNA strand exchange junction.
Plos One, 17, 2022
|
|
5WDO
 
 | | H-Ras bound to GMP-PNP at 277K | | Descriptor: | CALCIUM ION, GTPase HRas, MAGNESIUM ION, ... | | Authors: | Cofsky, J.C, Bandaru, P, Gee, C.L, Kuriyan, J. | | Deposit date: | 2017-07-05 | | Release date: | 2017-07-19 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Deconstruction of the Ras switching cycle through saturation mutagenesis.
Elife, 6, 2017
|
|
