8EXX
 
 | | Herpes simplex virus 1 polymerase holoenzyme bound to DNA and foscarnet (pre-translocation state) | | Descriptor: | DNA polymerase, DNA polymerase processivity factor, MAGNESIUM ION, ... | | Authors: | Pan, J, Abraham, J, Coen, D.M, Shankar, S, Yang, P, Hogle, J. | | Deposit date: | 2022-10-26 | | Release date: | 2024-09-04 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Viral DNA polymerase structures reveal mechanisms of antiviral drug resistance.
Cell, 187, 2024
|
|
5IWD
 
 | | HCMV DNA polymerase subunit UL44 complex with a small molecule | | Descriptor: | 5-methylidene-3-(methylsulfanyl)-2-benzothiophen-4(5H)-one, DNA polymerase processivity factor | | Authors: | Chen, H, Coen, D.M, Hogle, J.M, Filman, D.J. | | Deposit date: | 2016-03-22 | | Release date: | 2016-11-30 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | A Small Covalent Allosteric Inhibitor of Human Cytomegalovirus DNA Polymerase Subunit Interactions.
ACS Infect Dis, 3, 2017
|
|
5IXA
 
 | |
8V1Q
 
 | | Herpes simplex virus 1 polymerase holoenzyme bound to DNA in both open/closed conformations | | Descriptor: | DNA polymerase, DNA polymerase processivity factor, MAGNESIUM ION, ... | | Authors: | Pan, J, Abraham, J, Coen, D.M, Shankar, S, Yang, P, Hogle, J. | | Deposit date: | 2023-11-21 | | Release date: | 2024-09-04 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Viral DNA polymerase structures reveal mechanisms of antiviral drug resistance.
Cell, 187, 2024
|
|
8V1S
 
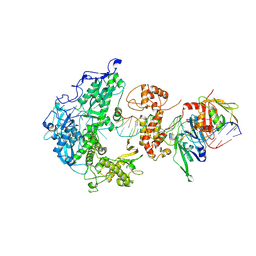 | | Herpes simplex virus 1 polymerase holoenzyme bound to mismatched DNA in editing conformation | | Descriptor: | DNA polymerase, DNA polymerase processivity factor, MAGNESIUM ION, ... | | Authors: | Pan, J, Abraham, J, Coen, D.M, Shankar, S, Yang, P, Hogle, J. | | Deposit date: | 2023-11-21 | | Release date: | 2024-09-04 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.26 Å) | | Cite: | Viral DNA polymerase structures reveal mechanisms of antiviral drug resistance.
Cell, 187, 2024
|
|
8V1T
 
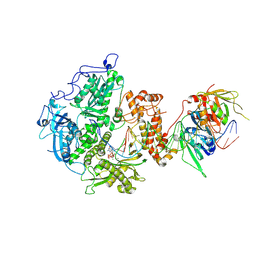 | | Herpes simplex virus 1 polymerase holoenzyme bound to DNA and acyclovir triphosphate in closed conformation | | Descriptor: | ACYCLOVIR TRIPHOSPHATE, DNA polymerase, DNA polymerase processivity factor, ... | | Authors: | Pan, J, Abraham, J, Coen, D.M, Shankar, S, Yang, P, Hogle, J. | | Deposit date: | 2023-11-21 | | Release date: | 2024-09-04 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Viral DNA polymerase structures reveal mechanisms of antiviral drug resistance.
Cell, 187, 2024
|
|
8V1R
 
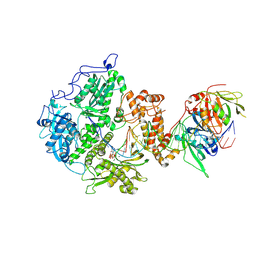 | | Herpes simplex virus 1 polymerase holoenzyme bound to DNA and DTTP in closed conformation | | Descriptor: | DNA polymerase, DNA polymerase processivity factor, MAGNESIUM ION, ... | | Authors: | Pan, J, Abraham, J, Coen, D.M, Shankar, S, Yang, P, Hogle, J. | | Deposit date: | 2023-11-21 | | Release date: | 2024-09-04 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Viral DNA polymerase structures reveal mechanisms of antiviral drug resistance.
Cell, 187, 2024
|
|
3I2M
 
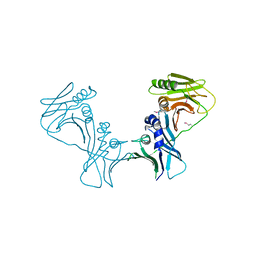 | | The Crystal Structure of PF-8, the DNA Polymerase Accessory Subunit from Kaposi s Sarcoma-Associated Herpesvirus | | Descriptor: | ORF59 | | Authors: | Baltz, J.L, Filman, D.J, Ciustea, M, Silverman, J.E.Y, Lautenschlager, C.L, Coen, D.M, Ricciardi, R.P, Hogle, J.M. | | Deposit date: | 2009-06-29 | | Release date: | 2010-05-12 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | The crystal structure of PF-8, the DNA polymerase accessory subunit from Kaposi's sarcoma-associated herpesvirus.
J.Virol., 83, 2009
|
|
3HSL
 
 | | The Crystal Structure of PF-8, the DNA Polymerase Accessory Subunit from Kaposi's Sarcoma-Associated Herpesvirus | | Descriptor: | ORF59 | | Authors: | Baltz, J.L, Filman, D.J, Ciustea, M, Silverman, J.E.Y, Lautenschlager, C.L, Coen, D.M, Ricciardi, R.P, Hogle, J.M. | | Deposit date: | 2009-06-10 | | Release date: | 2009-11-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The crystal structure of PF-8, the DNA polymerase accessory subunit from Kaposi's sarcoma-associated herpesvirus.
J.Virol., 83, 2009
|
|
1YYP
 
 | | Crystal structure of cytomegalovirus UL44 bound to C-terminal peptide from CMV UL54 | | Descriptor: | 1,2-ETHANEDIOL, DNA polymerase, DNA polymerase processivity factor, ... | | Authors: | Appleton, B.A, Brooks, J, Loregian, A, Filman, D.J, Coen, D.M, Hogle, J.M. | | Deposit date: | 2005-02-25 | | Release date: | 2005-12-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of the cytomegalovirus DNA polymerase subunit UL44 in complex with the C terminus from the catalytic subunit. Differences in structure and function relative to unliganded UL44.
J.Biol.Chem., 281, 2006
|
|
1DML
 
 | | CRYSTAL STRUCTURE OF HERPES SIMPLEX UL42 BOUND TO THE C-TERMINUS OF HSV POL | | Descriptor: | DNA POLYMERASE, DNA POLYMERASE PROCESSIVITY FACTOR | | Authors: | Zuccola, H.J, Filman, D.J, Coen, D.M, Hogle, J.M. | | Deposit date: | 1999-12-14 | | Release date: | 2000-03-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The crystal structure of an unusual processivity factor, herpes simplex virus UL42, bound to the C terminus of its cognate polymerase.
Mol.Cell, 5, 2000
|
|
1T6L
 
 | | Crystal Structure of the Human Cytomegalovirus DNA Polymerase Subunit, UL44 | | Descriptor: | DNA polymerase processivity factor | | Authors: | Appleton, B.A, Loregian, A, Filman, D.J, Coen, D.M, Hogle, J.M. | | Deposit date: | 2004-05-06 | | Release date: | 2004-08-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The Cytomegalovirus DNA Polymerase Subunit UL44 Forms a C Clamp-Shaped Dimer.
Mol.Cell, 15, 2004
|
|
5A3G
 
 | | Structure of herpesvirus nuclear egress complex subunit M50 | | Descriptor: | M50 | | Authors: | Leigh, K.E, Boeszoermenyi, A, Mansueto, M.S, Sharma, M, Filman, D.J, Coen, D.M, Wagner, G, Hogle, J.M, Arthanari, H. | | Deposit date: | 2015-06-01 | | Release date: | 2015-07-15 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Structure of a Herpesvirus Nuclear Egress Complex Subunit Reveals an Interaction Groove that is Essential for Viral Replication
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
