8ATU
 
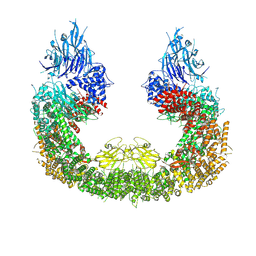 | | Cryo-EM structure of human BIRC6 | | Descriptor: | Baculoviral IAP repeat-containing protein 6, ZINC ION | | Authors: | Ehrmann, J.F, Grabarczyk, D.B, Clausen, T. | | Deposit date: | 2022-08-24 | | Release date: | 2023-02-15 | | Last modified: | 2023-03-29 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis for regulation of apoptosis and autophagy by the BIRC6/SMAC complex.
Science, 379, 2023
|
|
8AUK
 
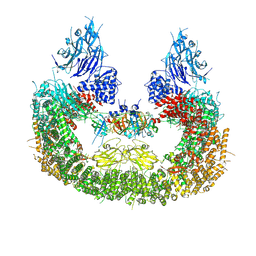 | | Cryo-EM structure of human BIRC6 in complex with HTRA2. | | Descriptor: | Baculoviral IAP repeat-containing protein 6, Serine protease HTRA2, mitochondrial, ... | | Authors: | Ehrmann, J.F, Grabarczyk, D.B, Clausen, T. | | Deposit date: | 2022-08-25 | | Release date: | 2023-02-15 | | Last modified: | 2023-05-24 | | Method: | ELECTRON MICROSCOPY (6.2 Å) | | Cite: | Structural basis for regulation of apoptosis and autophagy by the BIRC6/SMAC complex.
Science, 379, 2023
|
|
8B9O
 
 | |
8B9U
 
 | |
3CS0
 
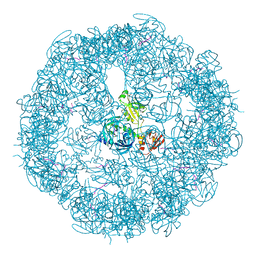 | | Crystal structure of DegP24 | | Descriptor: | Periplasmic serine endoprotease DegP, pentapeptide | | Authors: | Krojer, T, Sawa, J, Schaefer, E, Saibil, H.R, Ehrmann, M, Clausen, T. | | Deposit date: | 2008-04-08 | | Release date: | 2008-05-27 | | Last modified: | 2019-11-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis for the regulated protease and chaperone function of DegP
Nature, 453, 2008
|
|
7BII
 
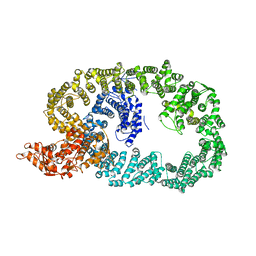 | | Crystal structure of Nematocida HUWE1 | | Descriptor: | E3 ubiquitin-protein ligase HUWE1 | | Authors: | Grabarczyk, D.B, Petrova, O.A, Meinhart, A, Kessler, D, Clausen, T. | | Deposit date: | 2021-01-12 | | Release date: | 2021-07-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.037 Å) | | Cite: | HUWE1 employs a giant substrate-binding ring to feed and regulate its HECT E3 domain.
Nat.Chem.Biol., 17, 2021
|
|
4GEQ
 
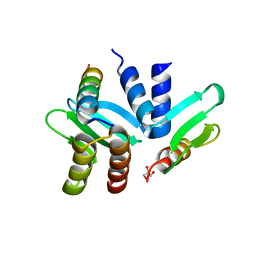 | | Crystal structure of the Spc24-Spc25/Cnn1 binding interface | | Descriptor: | GLYCEROL, Kinetochore protein SPC24, Kinetochore protein SPC25, ... | | Authors: | Malvezzi, F, Litos, G, Schleiffer, A, Heuck, A, Clausen, T, Westermann, S. | | Deposit date: | 2012-08-02 | | Release date: | 2013-01-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | A structural basis for kinetochore recruitment of the Ndc80 complex via two distinct centromere receptors.
Embo J., 32, 2013
|
|
5MZU
 
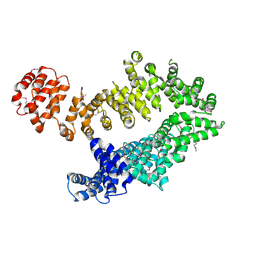 | |
1Z42
 
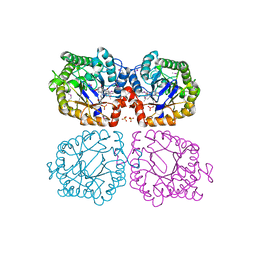 | | Crystal structure of oxidized YqjM from Bacillus subtilis complexed with p-hydroxybenzaldehyde | | Descriptor: | FLAVIN MONONUCLEOTIDE, P-HYDROXYBENZALDEHYDE, Probable NADH-dependent flavin oxidoreductase yqjM, ... | | Authors: | Kitzing, K, Fitzpatrick, T.B, Wilken, C, Sawa, J, Bourenkov, G.P, Macheroux, P, Clausen, T. | | Deposit date: | 2005-03-15 | | Release date: | 2005-05-17 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The 1.3 A Crystal Structure of the Flavoprotein YqjM Reveals a Novel Class of Old Yellow Enzymes
J.Biol.Chem., 280, 2005
|
|
1Z44
 
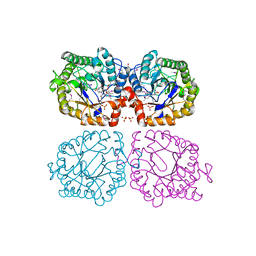 | | Crystal structure of oxidized YqjM from Bacillus subtilis complexed with p-nitrophenol | | Descriptor: | FLAVIN MONONUCLEOTIDE, P-NITROPHENOL, Probable NADH-dependent flavin oxidoreductase yqjM, ... | | Authors: | Kitzing, K, Fitzpatrick, T.B, Wilken, C, Sawa, J, Bourenkov, G.P, Macheroux, P, Clausen, T. | | Deposit date: | 2005-03-15 | | Release date: | 2005-05-17 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The 1.3 A Crystal Structure of the Flavoprotein YqjM Reveals a Novel Class of Old Yellow Enzymes
J.Biol.Chem., 280, 2005
|
|
4TRQ
 
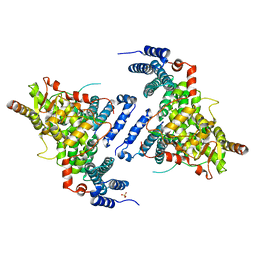 | | Crystal structure of Sac3/Thp1/Sem1 | | Descriptor: | 26S proteasome complex subunit SEM1, Nuclear mRNA export protein SAC3, Nuclear mRNA export protein THP1, ... | | Authors: | Hellerschmied, D, Schneider, S, Kohler, A, Clausen, T. | | Deposit date: | 2014-06-17 | | Release date: | 2015-08-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | The Nuclear Pore-Associated TREX-2 Complex Employs Mediator to Regulate Gene Expression.
Cell, 162, 2015
|
|
1Z41
 
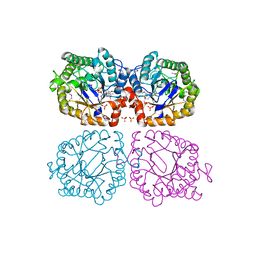 | | Crystal structure of oxidized YqjM from Bacillus subtilis | | Descriptor: | FLAVIN MONONUCLEOTIDE, Probable NADH-dependent flavin oxidoreductase yqjM, SULFATE ION | | Authors: | Kitzing, K, Fitzpatrick, T.B, Wilken, C, Sawa, J, Bourenkov, G.P, Macheroux, P, Clausen, T. | | Deposit date: | 2005-03-15 | | Release date: | 2005-05-17 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | The 1.3 A Crystal Structure of the Flavoprotein YqjM Reveals a Novel Class of Old Yellow Enzymes
J.Biol.Chem., 280, 2005
|
|
1Z48
 
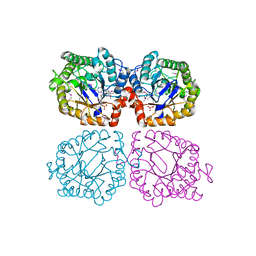 | | Crystal structure of reduced YqjM from Bacillus subtilis | | Descriptor: | FLAVIN MONONUCLEOTIDE, Probable NADH-dependent flavin oxidoreductase yqjM | | Authors: | Kitzing, K, Fitzpatrick, T.B, Wilken, C, Sawa, J, Bourenkov, G.P, Macheroux, P, Clausen, T. | | Deposit date: | 2005-03-15 | | Release date: | 2005-05-17 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The 1.3 A Crystal Structure of the Flavoprotein YqjM Reveals a Novel Class of Old Yellow Enzymes
J.Biol.Chem., 280, 2005
|
|
1N31
 
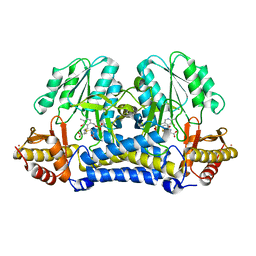 | | Structure of A Catalytically Inactive Mutant (K223A) of C-DES with a Substrate (Cystine) Linked to the Co-Factor | | Descriptor: | CYSTEINE, L-cysteine/cystine lyase C-DES, POTASSIUM ION, ... | | Authors: | Kaiser, J.T, Bruno, S, Clausen, T, Huber, R, Schiaretti, F, Mozzarelli, A, Kessler, D. | | Deposit date: | 2002-10-25 | | Release date: | 2003-01-21 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Snapshots of the Cystine Lyase "C-DES" during Catalysis: Studies in Solution and in the Crystalline State
J.Biol.Chem., 278, 2003
|
|
1N2T
 
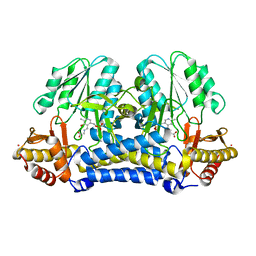 | | C-DES Mutant K223A with GLY Covalenty Linked to the PLP-cofactor | | Descriptor: | GLYCINE, L-cysteine/cystine lyase C-DES, POTASSIUM ION, ... | | Authors: | Kaiser, J.T, Bruno, S, Clausen, T, Huber, R, Schiaretti, F, Mozzarelli, A, Kessler, D. | | Deposit date: | 2002-10-24 | | Release date: | 2003-01-21 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Snapshots of the Cystine Lyase "C-DES" during Catalysis: Studies in Solution and in the Crystalline State
J.Biol.Chem., 278, 2003
|
|
4A9G
 
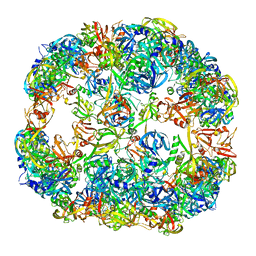 | | Symmetrized cryo-EM reconstruction of E. coli DegQ 24-mer in complex with beta-casein | | Descriptor: | PERIPLASMIC PH-DEPENDENT SERINE ENDOPROTEASE DEGQ | | Authors: | Malet, H, Canellas, F, Sawa, J, Yan, J, Thalassinos, K, Ehrmann, M, Clausen, T, Saibil, H.R. | | Deposit date: | 2011-11-26 | | Release date: | 2012-01-11 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (7.5 Å) | | Cite: | Newly Folded Substrates Inside the Molecular Cage of the Htra Chaperone Degq
Nat.Struct.Mol.Biol., 19, 2012
|
|
7OIK
 
 | |
7OIM
 
 | | Mouse RNF213, with mixed nucleotides bound | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, E3 ubiquitin-protein ligase RNF213, ... | | Authors: | Grabarczyk, D, Ahel, J, Clausen, T. | | Deposit date: | 2021-05-11 | | Release date: | 2022-06-01 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | E3 ubiquitin ligase RNF213 employs a non-canonical zinc finger active site and is allosterically regulated by ATP
To Be Published
|
|
6ZK7
 
 | | Crystal Structure of human PYROXD1/FAD complex | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Pyridine nucleotide-disulfide oxidoreductase domain-containing protein 1 | | Authors: | Meinhart, A, Asanovic, I, Martinez, J, Clausen, T. | | Deposit date: | 2020-06-30 | | Release date: | 2021-05-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | The oxidoreductase PYROXD1 uses NAD(P) + as an antioxidant to sustain tRNA ligase activity in pre-tRNA splicing and unfolded protein response.
Mol.Cell, 81, 2021
|
|
6QDJ
 
 | | Molecular features of the UNC-45 chaperone critical for binding and folding muscle myosin | | Descriptor: | 1,4-BUTANEDIOL, 2,5,8,11,14,17-HEXAOXANONADECAN-19-OL, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Meinhart, A, Clausen, T, Arnese, R. | | Deposit date: | 2019-01-02 | | Release date: | 2019-10-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.884 Å) | | Cite: | Molecular features of the UNC-45 chaperone critical for binding and folding muscle myosin.
Nat Commun, 10, 2019
|
|
6QDL
 
 | |
1VEU
 
 | | Crystal structure of the p14/MP1 complex at 2.15 A resolution | | Descriptor: | Late endosomal/lysosomal Mp1 interacting protein, Mitogen-activated protein kinase kinase 1 interacting protein 1 | | Authors: | Kurzbauer, R, Teis, D, Maurer-Stroh, S, Eisenhaber, F, Hekman, M, Bourenkov, G.P, Bartunik, H.D, Huber, L.A, Clausen, T. | | Deposit date: | 2004-04-05 | | Release date: | 2004-08-03 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structure of the p14/MP1 scaffolding complex: How a twin couple attaches mitogen- activated protein kinase signaling to late endosomes
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
6TV6
 
 | | Octameric McsB from Bacillus subtilis. | | Descriptor: | MAGNESIUM ION, Protein-arginine kinase | | Authors: | Suskiewicz, M.J, Hajdusits, B, Meinhart, A, Clausen, T. | | Deposit date: | 2020-01-09 | | Release date: | 2021-07-21 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | McsB forms a gated kinase chamber to mark aberrant bacterial proteins for degradation.
Elife, 10, 2021
|
|
1QGN
 
 | | CYSTATHIONINE GAMMA-SYNTHASE FROM NICOTIANA TABACUM | | Descriptor: | PROTEIN (CYSTATHIONINE GAMMA-SYNTHASE), PYRIDOXAL-5'-PHOSPHATE | | Authors: | Steegborn, C, Messerschmidt, A, Laber, B, Streber, W, Huber, R, Clausen, T. | | Deposit date: | 1999-05-02 | | Release date: | 1999-08-25 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The crystal structure of cystathionine gamma-synthase from Nicotiana tabacum reveals its substrate and reaction specificity.
J.Mol.Biol., 290, 1999
|
|
1N8P
 
 | | Crystal Structure of cystathionine gamma-lyase from yeast | | Descriptor: | Cystathionine gamma-lyase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Messerschmidt, A, Worbs, M, Steegborn, C, Wahl, M.C, Huber, R, Clausen, T. | | Deposit date: | 2002-11-21 | | Release date: | 2002-12-04 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Determinants of Enzymatic Specificity in the Cys-Met-Metabolism PLP-Dependent Enzymes Family: Crystal Structure of Cystathionine gamma-lyase from Yeast and Intrafamiliar Structural Comparison
BIOL.CHEM., 384, 2003
|
|
