7Q93
 
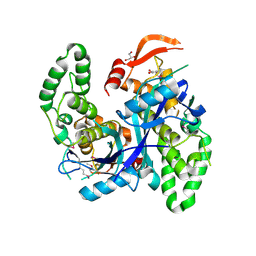 | | Crystal Structure of Agrobacterium tumefaciens NADQ, NAD complex. | | Descriptor: | GLYCEROL, NADQ transcription factor, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Cianci, M, Minazzato, G, Heroux, A, Raffaelli, N, Sorci, L, Gasparrini, M. | | Deposit date: | 2021-11-11 | | Release date: | 2022-11-23 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Bacterial NadQ (COG4111) is a Nudix-like, ATP-responsive regulator of NAD biosynthesis.
J.Struct.Biol., 214, 2022
|
|
7Q92
 
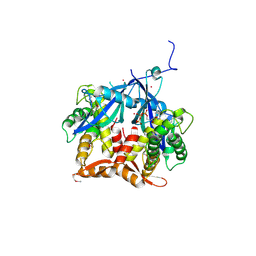 | | Crystal Structure of Agrobacterium tumefaciens NADQ, ATP complex. | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, NADQ transcription factor, POTASSIUM ION, ... | | Authors: | Cianci, M, Minazzato, G, Heroux, A, Raffaelli, N, Sorci, L, Gasparrini, M. | | Deposit date: | 2021-11-11 | | Release date: | 2022-11-09 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Bacterial NadQ (COG4111) is a Nudix-like, ATP-responsive regulator of NAD biosynthesis.
J.Struct.Biol., 214, 2022
|
|
6TP8
 
 | |
7PWY
 
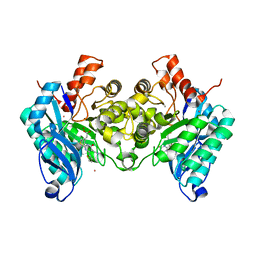 | | Structure of human dimeric ACMSD in complex with the inhibitor TES-1025 | | Descriptor: | 2-[3-[(5-cyano-6-oxidanylidene-4-thiophen-2-yl-1H-pyrimidin-2-yl)sulfanylmethyl]phenyl]ethanoic acid, 2-amino-3-carboxymuconate-6-semialdehyde decarboxylase, POTASSIUM ION, ... | | Authors: | Cianci, M, Giacche, N, Carotti, A, Liscio, P, Amici, A, Cialabrini, L, De Franco, F, Pellicciari, R, Raffaelli, N. | | Deposit date: | 2021-10-07 | | Release date: | 2022-04-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Basis of Human Dimeric alpha-Amino-beta-Carboxymuconate-epsilon-Semialdehyde Decarboxylase Inhibition With TES-1025.
Front Mol Biosci, 9, 2022
|
|
1H91
 
 | | The crystal structure of lobster apocrustacyanin A1 using softer X-rays. | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CRUSTACYANIN A1 SUBUNIT | | Authors: | Cianci, M, Rizkallah, P.J, Olczak, A, Raftery, J, Chayen, N.E, Zagalsky, P.F, Helliwell, J.R. | | Deposit date: | 2001-02-21 | | Release date: | 2001-09-06 | | Last modified: | 2019-05-22 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure of Lobster Apocrustacyanin A1 Using Softer X-Rays
Acta Crystallogr.,Sect.D, 57, 2001
|
|
6H2M
 
 | | Long wavelength Mesh&Collect native SAD phasing on microcrystals | | Descriptor: | CALCIUM ION, Concanavalin-A,Concanavalin-A, MANGANESE (II) ION | | Authors: | Cianci, M, Nanao, M, Schneider, T.R. | | Deposit date: | 2018-07-13 | | Release date: | 2019-03-13 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.929 Å) | | Cite: | Long-wavelength Mesh&Collect native SAD phasing from microcrystals.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
5FTP
 
 | | sulfur SAD phasing of Cdc23Nterm: data collection with a tailored X- ray beam size at 2.69 A wavelength (4.6 keV) | | Descriptor: | ANAPHASE-PROMOTING COMPLEX SUBUNIT 8 | | Authors: | Cianci, M, Groves, M.R, Barford, D, Schneider, T.R. | | Deposit date: | 2016-01-14 | | Release date: | 2016-03-16 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Data Collection with a Tailored X-Ray Beam Size at 2.69 A Wavelength (4.6 Kev): Sulfur Sad Phasing of Cdc23Nterm
Acta Crystallogr.,Sect.D, 72, 2016
|
|
7Q94
 
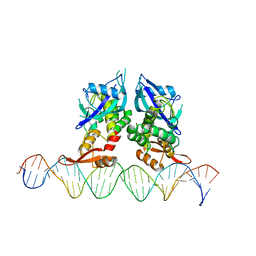 | | Crystal Structure of Agrobacterium tumefaciens NADQ, DNA complex. | | Descriptor: | DNA binding region (31-MER), NADQ transcription factor | | Authors: | Cianci, M, Minazzato, G, Heroux, A, Raffaelli, N, Sorci, L, Gasparrini, M. | | Deposit date: | 2021-11-11 | | Release date: | 2022-11-09 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (4.3 Å) | | Cite: | Bacterial NadQ (COG4111) is a Nudix-like, ATP-responsive regulator of NAD biosynthesis.
J.Struct.Biol., 214, 2022
|
|
7Q91
 
 | | Crystal Structure of Agrobacterium tumefaciens NADQ, native form. | | Descriptor: | NADQ transcription factor, SODIUM ION | | Authors: | Cianci, M, Minazzato, G, Heroux, A, Raffaelli, N, Sorci, L, Gasparrini, M. | | Deposit date: | 2021-11-11 | | Release date: | 2022-11-09 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Bacterial NadQ (COG4111) is a Nudix-like, ATP-responsive regulator of NAD biosynthesis.
J.Struct.Biol., 214, 2022
|
|
1GKA
 
 | | The molecular basis of the coloration mechanism in lobster shell. beta-crustacyanin at 3.2 A resolution | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ASTAXANTHIN, ... | | Authors: | Cianci, M, Rizkallah, P.J, Olczak, A, Raftery, J, Chayen, N.E, Zagalsky, P.F, Helliwell, J.R. | | Deposit date: | 2001-08-10 | | Release date: | 2002-08-08 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.23 Å) | | Cite: | The Molecular Basis of the Coloration Mechanism in Lobster Shell: Beta -Crustacyanin at 3.2-A Resolution
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
2WUV
 
 | | Crystallographic analysis of counter-ion effects on subtilisin enzymatic action in acetonitrile | | Descriptor: | ACETONITRILE, CALCIUM ION, CESIUM ION, ... | | Authors: | Cianci, M, Tomaszewki, B, Helliwell, J.R, Halling, P.J. | | Deposit date: | 2009-10-09 | | Release date: | 2010-12-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Crystallographic Analysis of Counterion Effects on Subtilisin Enzymatic Action in Acetonitrile.
J.Am.Chem.Soc., 132, 2010
|
|
2WUW
 
 | | Crystallographic analysis of counter-ion effects on subtilisin enzymatic action in acetonitrile (native data) | | Descriptor: | ACETONITRILE, CALCIUM ION, SODIUM ION, ... | | Authors: | Cianci, M, Tomaszewki, B, Helliwell, J.R, Halling, P.J. | | Deposit date: | 2009-10-09 | | Release date: | 2010-12-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Crystallographic Analysis of Counterion Effects on Subtilisin Enzymatic Action in Acetonitrile.
J.Am.Chem.Soc., 132, 2010
|
|
2W1M
 
 | | THE INTERDEPENDENCE OF WAVELENGTH, REDUNDANCY AND DOSE IN SULFUR SAD EXPERIMENTS: 2.070 A WAVELENGTH with 2theta 30 degrees data | | Descriptor: | CHLORIDE ION, LYSOZYME C, SODIUM ION | | Authors: | Cianci, M, Helliwell, J.R, Suzuki, A. | | Deposit date: | 2008-10-17 | | Release date: | 2008-11-04 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | The Interdependence of Wavelength, Redundancy and Dose in Sulfur Sad Experiments.
Acta Crystallogr.,Sect.D, 64, 2008
|
|
2W1Y
 
 | | THE INTERDEPENDENCE OF WAVELENGTH, REDUNDANCY AND DOSE IN SULFUR SAD EXPERIMENTS: 1.540 A wavelength 180 images data | | Descriptor: | CHLORIDE ION, LYSOZYME C, SODIUM ION | | Authors: | Cianci, M, Helliwell, J.R, Suzuki, A. | | Deposit date: | 2008-10-21 | | Release date: | 2008-11-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | The Interdependence of Wavelength, Redundancy and Dose in Sulfur Sad Experiments.
Acta Crystallogr.,Sect.D, 64, 2008
|
|
2W1L
 
 | | THE INTERDEPENDENCE OF WAVELENGTH, REDUNDANCY AND DOSE IN SULFUR SAD EXPERIMENTS: 0.979 a wavelength 991 images data | | Descriptor: | CHLORIDE ION, LYSOZYME C, SODIUM ION | | Authors: | Cianci, M, Helliwell, J.R, Suzuki, A. | | Deposit date: | 2008-10-17 | | Release date: | 2008-10-28 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | The Interdependence of Wavelength, Redundancy and Dose in Sulfur Sad Experiments.
Acta Crystallogr.,Sect.D, 64, 2008
|
|
4C3U
 
 | | Extensive counter-ion interactions with subtilisin in aqueous medium, Cs derivative | | Descriptor: | CALCIUM ION, CESIUM ION, CHLORIDE ION, ... | | Authors: | Cianci, M, Negroni, J, Helliwell, J.R, Halling, P.J. | | Deposit date: | 2013-08-27 | | Release date: | 2014-08-20 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Extensive Counter-Ion Interactions Seen at the Surface of Subtilisin in an Aqueous Medium
Rsc Adv, 2014
|
|
2W1X
 
 | | The interdependence of wavelength, redundancy and dose in sulfur SAD experiments: 1.284 A wavelength 360 images data | | Descriptor: | CHLORIDE ION, LYSOZYME C, SODIUM ION | | Authors: | Cianci, M, Helliwell, J.R, Suzuki, A. | | Deposit date: | 2008-10-21 | | Release date: | 2008-11-04 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Interdependence of Wavelength, Redundancy and Dose in Sulfur Sad Experiments.
Acta Crystallogr.,Sect.D, 64, 2008
|
|
4C3V
 
 | | Extensive counter-ion interactions with subtilisin in aqueous medium, no Cs soak | | Descriptor: | CALCIUM ION, SODIUM ION, SUBTILISIN CARLSBERG, ... | | Authors: | Cianci, M, Negroni, J, Helliwell, J.R, Halling, P.J. | | Deposit date: | 2013-08-27 | | Release date: | 2014-08-20 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Extensive Counter-Ion Interactions Seen at the Surface of Subtilisin in an Aqueous Medium
Rsc Adv, 2014
|
|
5AHS
 
 | | 3-Sulfinopropionyl-Coenzyme A (3SP-CoA) desulfinase from Advenella mimgardefordensis DPN7T: holo crystal structure with the substrate analog succinyl-CoA | | Descriptor: | ACYL-COA DEHYDROGENASE, COENZYME A, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Cianci, M, Schuermann, M, Meijers, R, Schneider, T.R, Steinbuechel, A. | | Deposit date: | 2015-02-06 | | Release date: | 2015-06-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | 3-Sulfinopropionyl-Coenzyme a (3Sp-Coa) Desulfinase from Advenella Mimigardefordensis Dpn7(T): Crystal Structure and Function of a Desulfinase with an Acyl-Coa Dehydrogenase Fold.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
5AF7
 
 | | 3-Sulfinopropionyl-coenzyme A (3SP-CoA) desulfinase from Advenella mimigardefordensis DPN7T: crystal structure and function of a desulfinase with an acyl-CoA dehydrogenase fold. Native crystal structure | | Descriptor: | ACYL-COA DEHYDROGENASE, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL | | Authors: | Cianci, M, Schuermann, M, Meijers, R, Schneider, T.R, Steinbuechel, A. | | Deposit date: | 2015-01-20 | | Release date: | 2015-06-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | 3-Sulfinopropionyl-Coenzyme a (3Sp-Coa) Desulfinase from Advenella Mimigardefordensis Dpn7(T): Crystal Structure and Function of a Desulfinase with an Acyl-Coa Dehydrogenase Fold.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
6ZO1
 
 | | 1.61 A resolution 3,5-dimethylcatechol (3,5-dimethylbenzene-1,2-diol) inhibited Sporosarcina pasteurii urease | | Descriptor: | 1,2-ETHANEDIOL, HYDROXIDE ION, NICKEL (II) ION, ... | | Authors: | Mazzei, L, Cianci, M, Musiani, F, Ciurli, S. | | Deposit date: | 2020-07-07 | | Release date: | 2020-12-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Inhibition of Urease, a Ni-Enzyme: The Reactivity of a Key Thiol With Mono- and Di-Substituted Catechols Elucidated by Kinetic, Structural, and Theoretical Studies.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
2B5S
 
 | | Crystal structure of peach Pru p3, the prototypic member of the family of plant non-specific lipid transfer protein pan-allergens | | Descriptor: | HEPTANE, LAURIC ACID, Non-specific lipid transfer protein, ... | | Authors: | Pasquato, N, Berni, R, Folli, C, Folloni, S, Cianci, M, Pantano, S, Helliwell, R.J, Zanotti, G. | | Deposit date: | 2005-09-29 | | Release date: | 2005-11-29 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal Structure of Peach Pru p 3, the Prototypic Member of the Family of Plant Non-specific Lipid Transfer Protein Pan-allergens
J.Mol.Biol., 356, 2006
|
|
4AC7
 
 | | The crystal structure of Sporosarcina pasteurii urease in complex with citrate | | Descriptor: | 1,2-ETHANEDIOL, CITRATE ANION, HYDROXIDE ION, ... | | Authors: | Benini, S, Kosikowska, P, Cianci, M, Gonzalez Vara, A, Berlicki, L, Ciurli, S. | | Deposit date: | 2011-12-14 | | Release date: | 2013-01-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The Crystal Structure of Sporosarcina Pasteurii Urease in a Complex with Citrate Provides New Hints for Inhibitor Design.
J.Biol.Inorg.Chem., 18, 2013
|
|
8C86
 
 | | Crystal structure of human transthyretin in complex with 3-O-methyltolcapone analogue 2 | | Descriptor: | (2,4-dimethylphenyl)-(3-methoxy-5-nitro-4-oxidanyl-phenyl)methanone, Transthyretin | | Authors: | Poonsiri, T, Benini, S, Loconte, V, Cianci, M. | | Deposit date: | 2023-01-18 | | Release date: | 2024-01-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | 3-O-Methyltolcapone and Its Lipophilic Analogues Are Potent Inhibitors of Transthyretin Amyloidogenesis with High Permeability and Low Toxicity.
Int J Mol Sci, 25, 2023
|
|
8C85
 
 | | Crystal structure of human transthyretin in complex with 3-O-methyltolcapone analogue 1 | | Descriptor: | (3,5-dimethylphenyl)-(3-methoxy-5-nitro-4-oxidanyl-phenyl)methanone, Transthyretin | | Authors: | Poonsiri, T, Benini, S, Loconte, V, Cianci, M. | | Deposit date: | 2023-01-18 | | Release date: | 2024-01-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.19 Å) | | Cite: | 3-O-Methyltolcapone and Its Lipophilic Analogues Are Potent Inhibitors of Transthyretin Amyloidogenesis with High Permeability and Low Toxicity.
Int J Mol Sci, 25, 2023
|
|
