5GIK
 
 | | Modulation of the affinity of a HIV-1 capsid-directed ankyrin towards its viral target through critical amino acid editing | | Descriptor: | Artificial ankyrin repeat protein_Ank(GAG)1D4 mutant -S45Y, GLYCEROL | | Authors: | Chuankhayan, P, Saoin, S, Wisitponchai, T, Intachai, K, Chupradit, K, Moonmuang, S, Kitidee, K, Nangola, S, Sanghiran, L.V, Hong, S.S, Boulanger, P, Tayapiwatana, C, Chen, C.J. | | Deposit date: | 2016-06-24 | | Release date: | 2017-06-28 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Modulation of the affinity of a HIV-1 capsid-directed ankyrintowards its viral target through critical amino acid editing
To Be Published
|
|
4HLL
 
 | | Crystal structure of Artificial ankyrin repeat protein_Ank(GAG)1D4 | | Descriptor: | Ankyrin(GAG)1D4 | | Authors: | Chuankhayan, P, Nangola, S, Minard, P, Boulanger, P, Hong, S.S, Tayapiwatana, C, Chen, C.-J. | | Deposit date: | 2012-10-17 | | Release date: | 2013-10-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Identification of Gag bioactive determinants specific to designed ankyrin and interfering in HIV-1 assembly
To be Published
|
|
4ZFH
 
 | | Crystal structure of Artificial ankyrin repeat protein_Ank(GAG)1D4 mutant -Y56A | | Descriptor: | Artificial ankyrin repeat protein_Ank(GAG)1D4 mutant -Y56A, MAGNESIUM ION | | Authors: | Chuankhayan, P, Saoin, S, Chupradit, K, Wisitponchai, T, Intachai, K, Kitidee, K, Nangola, S, Sanghiran, L.V, Hong, S.S, Boulanger, P, Tayapiwatana, C, Chen, C.J. | | Deposit date: | 2015-04-21 | | Release date: | 2016-04-20 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Crystal structure of Artificial ankyrin repeat protein_Ank(GAG)1D4 mutant- Y56A
To Be Published
|
|
4XPX
 
 | | Crystal structure of hemerythrin:wild-type | | Descriptor: | Bacteriohemerythrin, FE (II) ION | | Authors: | Chuankhayan, P, Chen, K.H.C, Wu, H.H, Chen, C.J, Fukuda, M, Yu, S.S.F, Chan, S.I. | | Deposit date: | 2015-01-18 | | Release date: | 2015-04-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.03 Å) | | Cite: | The bacteriohemerythrin from Methylococcus capsulatus (Bath): Crystal structures reveal that Leu114 regulates a water tunnel.
J.Inorg.Biochem., 150, 2015
|
|
4XQ1
 
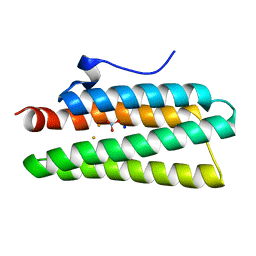 | | Crystal structure of hemerythrin: L114A mutant | | Descriptor: | Bacteriohemerythrin, FE (III) ION, NITRATE ION, ... | | Authors: | Chuankhayan, P, Chen, K.H.C, Wu, H.H, Chen, C.J, Fukuda, M, Yu, S.S.F, Chan, S.I. | | Deposit date: | 2015-01-18 | | Release date: | 2015-04-29 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The bacteriohemerythrin from Methylococcus capsulatus (Bath): Crystal structures reveal that Leu114 regulates a water tunnel.
J.Inorg.Biochem., 150, 2015
|
|
4XPW
 
 | | Crystal structures of Leu114F mutant | | Descriptor: | Bacteriohemerythrin, FE (II) ION, GLYCEROL | | Authors: | Chuankhayan, P, Chen, K.H.C, Wu, H.H, Chen, C.J, Fukuda, M, Yu, S.S.F, Chan, S.I. | | Deposit date: | 2015-01-18 | | Release date: | 2015-04-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.17 Å) | | Cite: | The bacteriohemerythrin from Methylococcus capsulatus (Bath): Crystal structures reveal that Leu114 regulates a water tunnel.
J.Inorg.Biochem., 150, 2015
|
|
4XPY
 
 | | Crystal structure of hemerythrin : L114Y mutant | | Descriptor: | Bacteriohemerythrin, FE (II) ION, GLYCEROL | | Authors: | Chuankhayan, P, Chen, K.H.C, Wu, H.H, Chen, C.J, Fukuda, M, Yu, S.S.F, Chan, S.I. | | Deposit date: | 2015-01-18 | | Release date: | 2015-04-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.13 Å) | | Cite: | The bacteriohemerythrin from Methylococcus capsulatus (Bath): Crystal structures reveal that Leu114 regulates a water tunnel.
J.Inorg.Biochem., 150, 2015
|
|
4L8W
 
 | | Crystal structure of gamma glutamyl hydrolase (H218N) from zebrafish complex with MTX polyglutamate | | Descriptor: | D-GLUTAMIC ACID, Gamma-glutamyl hydrolase, METHOTREXATE | | Authors: | Chuankhayan, P, Kao, T.-T, Chen, C.-J, Fu, T.-F. | | Deposit date: | 2013-06-18 | | Release date: | 2014-05-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Structural insights into the hydrolysis and polymorphism of methotrexate polyglutamate by zebrafish gamma-glutamyl hydrolase
J.Med.Chem., 56, 2013
|
|
4L8Y
 
 | |
4L8F
 
 | | Crystal structure of gamma-glutamyl hydrolase (C108A) complex with MTX | | Descriptor: | Gamma-glutamyl hydrolase, METHOTREXATE | | Authors: | Chuankhayan, P, Kao, T.-T, Chen, C.-J, Fu, T.-F. | | Deposit date: | 2013-06-17 | | Release date: | 2014-05-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structural insights into the hydrolysis and polymorphism of methotrexate polyglutamate by zebrafish gamma-glutamyl hydrolase
J.Med.Chem., 56, 2013
|
|
4L7Q
 
 | | Crystal structure of gamma glutamyl hydrolase (wild-type) from zebrafish | | Descriptor: | GLYCEROL, Gamma-glutamyl hydrolase | | Authors: | Chuankhayan, P, Kao, T.-T, Chen, C.-J, Fu, T.-F. | | Deposit date: | 2013-06-14 | | Release date: | 2014-05-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural insights into the hydrolysis and polymorphism of methotrexate polyglutamate by zebrafish gamma-glutamyl hydrolase
J.Med.Chem., 56, 2013
|
|
4L95
 
 | | Crystal structure of gamma glutamyl hydrolase (H218N) from zebrafish | | Descriptor: | GLYCEROL, Gamma-glutamyl hydrolase | | Authors: | Chuankhayan, P, Kao, T.-T, Chen, C.-J, Fu, T.-F. | | Deposit date: | 2013-06-18 | | Release date: | 2014-05-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Structural insights into the hydrolysis and polymorphism of methotrexate polyglutamate by zebrafish gamma-glutamyl hydrolase
J.Med.Chem., 56, 2013
|
|
3LF7
 
 | |
3LDK
 
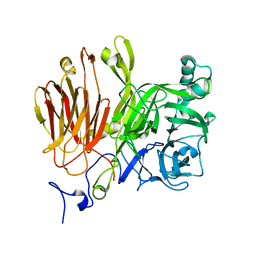 | | Crystal Structure of A. japonicus CB05 | | Descriptor: | Fructosyltransferase, beta-D-fructofuranose-(2-1)-alpha-D-glucopyranose | | Authors: | Chuankhayan, P, Chen, C.J, Chaing, C.M, Hsieh, C.Y, Chen, C.D, Hsieh, Y.C. | | Deposit date: | 2010-01-13 | | Release date: | 2010-05-12 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of Aspergillus japonicus fructosyltransferase complex with donor/acceptor substrates reveal complete sbusites in the active site for catalysis
To be Published
|
|
3LIH
 
 | |
3LIG
 
 | |
3LEM
 
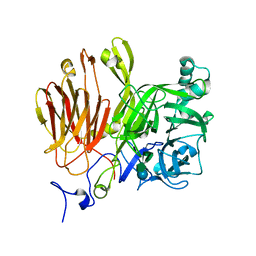 | |
3LDR
 
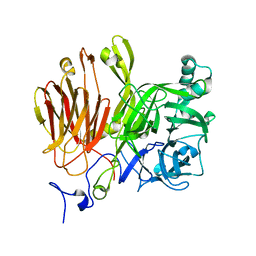 | |
3LFI
 
 | |
7CDL
 
 | | holo-methanol dehydrogenase (MDH) with Cys131-Cys132 reduced from Methylococcus capsulatus (Bath) | | Descriptor: | CALCIUM ION, Methanol dehydrogenase [cytochrome c] subunit 2, Methanol dehydrogenase protein, ... | | Authors: | Chuankhayan, P, Chan, S.I, Nareddy, P.K.R, Tsai, I.K, Tsai, Y.F, Chen, K.H.-C, Yu, S.S.-F, Chen, C.J. | | Deposit date: | 2020-06-20 | | Release date: | 2021-06-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Mechanism of Pyrroloquinoline Quinone-Dependent Hydride Transfer Chemistry from Spectroscopic and High-Resolution X-ray Structural Studies of the Methanol Dehydrogenase from Methylococcus capsulatus (Bath).
J.Am.Chem.Soc., 143, 2021
|
|
7CE9
 
 | | PQQ-soaked Apo-methanol dehydrogenase (MDH) from Methylococcus capsulatus (Bath) | | Descriptor: | CALCIUM ION, Methanol dehydrogenase [cytochrome c] subunit 2, Methanol dehydrogenase protein, ... | | Authors: | Chuankhayan, P, Chan, S.I, Nareddy, P.K.R, Tsai, I.K, Tsai, Y.F, Chen, K.H.-C, Yu, S.S.-F, Chen, C.J. | | Deposit date: | 2020-06-22 | | Release date: | 2021-06-30 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Mechanism of Pyrroloquinoline Quinone-Dependent Hydride Transfer Chemistry from Spectroscopic and High-Resolution X-ray Structural Studies of the Methanol Dehydrogenase from Methylococcus capsulatus (Bath).
J.Am.Chem.Soc., 143, 2021
|
|
7CFX
 
 | | NAD-soaked Holo-methanol dehydrogenase (MDH) from Methylococcus capsulatus (Bath) | | Descriptor: | CALCIUM ION, Methanol dehydrogenase [cytochrome c] subunit 2, Methanol dehydrogenase protein, ... | | Authors: | Chuankhayan, P, Chan, S.I, Nareddy, P.K.R, Tsai, I.K, Tsai, Y.F, Chen, K.H.-C, Yu, S.S.-F, Chen, C.J. | | Deposit date: | 2020-06-29 | | Release date: | 2021-06-30 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Mechanism of Pyrroloquinoline Quinone-Dependent Hydride Transfer Chemistry from Spectroscopic and High-Resolution X-ray Structural Studies of the Methanol Dehydrogenase from Methylococcus capsulatus (Bath).
J.Am.Chem.Soc., 143, 2021
|
|
7CED
 
 | | Apo-methanol dehydrogenase (MDH) from Methylococcus capsulatus (Bath) | | Descriptor: | Methanol dehydrogenase [cytochrome c] subunit 2, Methanol dehydrogenase protein, large subunit | | Authors: | Chuankhayan, P, Chan, S.I, Nareddy, P.K.R, Tsai, I.K, Tsai, Y.F, Chen, K.H.-C, Yu, S.S.-F, Chen, C.J. | | Deposit date: | 2020-06-22 | | Release date: | 2021-06-30 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Mechanism of Pyrroloquinoline Quinone-Dependent Hydride Transfer Chemistry from Spectroscopic and High-Resolution X-ray Structural Studies of the Methanol Dehydrogenase from Methylococcus capsulatus (Bath).
J.Am.Chem.Soc., 143, 2021
|
|
7CE5
 
 | | Methanol-PQQ bound methanol dehydrogenase (MDH) from Methylococcus capsulatus (Bath) | | Descriptor: | CALCIUM ION, METHANOL, Methanol dehydrogenase [cytochrome c] subunit 2, ... | | Authors: | Chuankhayan, P, Chan, S.I, Nareddy, P.K.R, Tsai, I.K, Tsai, Y.F, Chen, K.H.-C, Yu, S.S.-F, Chen, C.J. | | Deposit date: | 2020-06-22 | | Release date: | 2021-06-30 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Mechanism of Pyrroloquinoline Quinone-Dependent Hydride Transfer Chemistry from Spectroscopic and High-Resolution X-ray Structural Studies of the Methanol Dehydrogenase from Methylococcus capsulatus (Bath).
J.Am.Chem.Soc., 143, 2021
|
|
7EXH
 
 | | Crystal structure of D383A mutant from Arabidopsis thaliana complexed with Galactinol. | | Descriptor: | Probable galactinol--sucrose galactosyltransferase 6, galactinol | | Authors: | Chuankhayan, P, Guan, H.H, Lin, C.C, Chen, N.C, Huang, Y.C, Yoshimura, M, Nakagawa, A, Lee, R.H, Chen, C.J. | | Deposit date: | 2021-05-27 | | Release date: | 2022-11-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.63 Å) | | Cite: | Structural insight into the hydrolase and synthase activities of an alkaline alpha-galactosidase from Arabidopsis from complexes with substrate/product.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
