8CDF
 
 | |
7AY3
 
 | |
6F7J
 
 | |
6F8D
 
 | |
3X2S
 
 | | Crystal structure of pyrene-conjugated adenylate kinase | | Descriptor: | Adenylate kinase, BIS(ADENOSINE)-5'-PENTAPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Fujii, A, Sekiguchi, Y, Matsumura, H, Inoue, T, Chung, W.-S, Hirota, S, Matsuo, T. | | Deposit date: | 2014-12-31 | | Release date: | 2015-04-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Excimer Emission Properties on Pyrene-Labeled Protein Surface: Correlation between Emission Spectra, Ring Stacking Modes, and Flexibilities of Pyrene Probes.
Bioconjug.Chem., 26, 2015
|
|
5EUN
 
 | | The Crystal Structure of Human Kynurenine Aminotransferase II, PLP-bound form, at 1.83 A | | Descriptor: | Kynurenine/alpha-aminoadipate aminotransferase, mitochondrial | | Authors: | Nematollahi, A, Sun, G, Kwan, A, Jeffries, C.M, Harrop, S.J, Hanrahan, J.R, Nadvi, N.A, Church, W.B. | | Deposit date: | 2015-11-18 | | Release date: | 2015-12-16 | | Last modified: | 2016-08-10 | | Method: | X-RAY DIFFRACTION (1.825 Å) | | Cite: | Structure of the PLP-Form of the Human Kynurenine Aminotransferase II in a Novel Spacegroup at 1.83 angstrom Resolution.
Int J Mol Sci, 17, 2016
|
|
5EFS
 
 | | The crystal structure of human kynurenine aminotransferase II | | Descriptor: | Kynurenine/alpha-aminoadipate aminotransferase, mitochondrial | | Authors: | Nematollahi, A, Sun, G, Kwan, A, Harrop, S.J, Hanrahan, J.R, Nadvi, N.A, Church, W.B. | | Deposit date: | 2015-10-26 | | Release date: | 2015-11-11 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.82503 Å) | | Cite: | The crystal structure of human kynurenine aminotransferase II
To Be Published
|
|
6D0A
 
 | |
4CC2
 
 | | Complex of human Tuba C-terminal SH3 domain with human N-WASP proline- rich peptide - P212121 | | Descriptor: | CHLORIDE ION, DYNAMIN-BINDING PROTEIN, GLYCEROL, ... | | Authors: | Polle, L, Rigano, L, Julian, R, Ireton, K, Schubert, W.-D. | | Deposit date: | 2013-10-17 | | Release date: | 2013-10-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural Details of Human Tuba Recruitment by Inlc of Listeria Monocytogenes Elucidate Bacterial Cell-Cell Spreading.
Structure, 22, 2014
|
|
4CC3
 
 | | Complex of human Tuba C-terminal SH3 domain and Mena proline-rich peptide - H3 | | Descriptor: | 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, CHLORIDE ION, DYNAMIN-BINDING PROTEIN, ... | | Authors: | Polle, L, Rigano, L, Julian, R, Ireton, K, Schubert, W.-D. | | Deposit date: | 2013-10-17 | | Release date: | 2013-10-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structural Details of Human Tuba Recruitment by Inlc of Listeria Monocytogenes Elucidate Bacterial Cell-Cell Spreading.
Structure, 22, 2014
|
|
4COK
 
 | | Functional and Structural Characterization of Pyruvate decarboxylase from Gluconoacetobacter diazotrophicus | | Descriptor: | MAGNESIUM ION, PYRUVATE DECARBOXYLASE, SULFATE ION, ... | | Authors: | vanZyl, L.J, Schubert, W.-D, Tuffin, M, Cowan, D.A. | | Deposit date: | 2014-01-29 | | Release date: | 2014-10-01 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Structure and Functional Characterization of Pyruvate Decarboxylase from Gluconacetobacter Diazotrophicus.
Bmc Struct.Biol., 14, 2014
|
|
7YPL
 
 | |
4LXO
 
 | | Crystal structure of 9,10Fn3-elegantin chimera | | Descriptor: | CALCIUM ION, Fibronectin | | Authors: | Chang, Y.S, Shiu, J.H, Chuang, W.J. | | Deposit date: | 2013-07-30 | | Release date: | 2014-08-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Design, Structure Determination, and Biological Evaluation of Potent Integrin Alpha5beta1-Specific Antagonist Using the Ninth and Tenth Module of Fibronectin Type III Domain
To be Published
|
|
7ZSZ
 
 | |
2C6Y
 
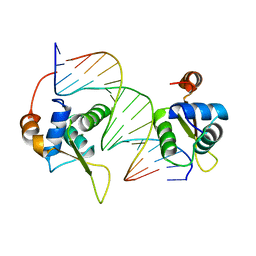 | | Crystal structure of interleukin enhancer-binding factor 1 bound to DNA | | Descriptor: | FORKHEAD BOX PROTEIN K2, INTERLEUKIN 2 PROMOTOR, MAGNESIUM ION | | Authors: | Tsai, K.-L, Huang, C.-Y, Chang, C.-H, Sun, Y.-J, Chuang, W.-J, Hsiao, C.-D. | | Deposit date: | 2005-11-15 | | Release date: | 2006-04-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of the Human Foxk1A-DNA Complex and its Implications on the Diverse Binding Specificity of Winged Helix/Forkhead Proteins.
J.Biol.Chem., 281, 2006
|
|
1JZU
 
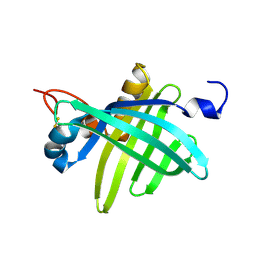 | | Cell transformation by the myc oncogene activates expression of a lipocalin: analysis of the gene (Q83) and solution structure of its protein product | | Descriptor: | lipocalin Q83 | | Authors: | Hartl, M, Matt, T, Schueler, W, Siemeister, G, Kontaxis, G, Kloiber, K, Konrat, R, Bister, K. | | Deposit date: | 2001-09-17 | | Release date: | 2003-07-15 | | Last modified: | 2021-10-27 | | Method: | SOLUTION NMR | | Cite: | Cell Transformation by the v-myc Oncogene Abrogates c-Myc/Max-mediated Suppression of a
C/EBPbeta-dependent Lipocalin Gene.
J.Mol.Biol., 333, 2003
|
|
2N3M
 
 | |
1IQB
 
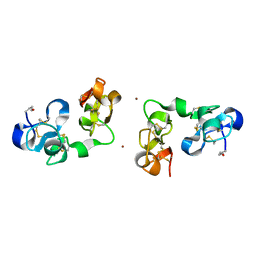 | |
4LMY
 
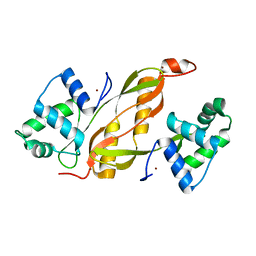 | | Structure of GAS PerR-Zn-Zn | | Descriptor: | Peroxide stress regulator PerR, FUR family, ZINC ION | | Authors: | Lin, C.S, Chao, S.Y, Nix, J.C, Tseng, H.L, Tsou, C.C, Fei, C.H, Ciou, H.S, Jeng, U.S, Lin, Y.S, Chuang, W.J, Wu, J.J, Wang, S. | | Deposit date: | 2013-07-11 | | Release date: | 2014-04-02 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Distinct structural features of the peroxide response regulator from group a streptococcus drive DNA binding
Plos One, 9, 2014
|
|
7X4S
 
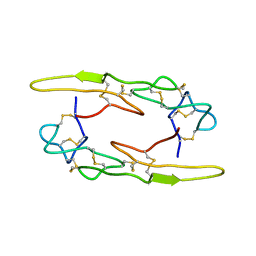 | |
7X4V
 
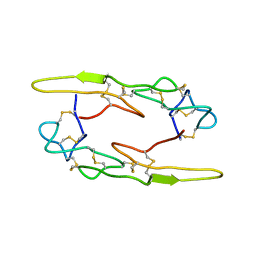 | |
7X4Z
 
 | |
1GPJ
 
 | | Glutamyl-tRNA Reductase from Methanopyrus kandleri | | Descriptor: | (2R,3R,4S,5S)-4-AMINO-2-[6-(DIMETHYLAMINO)-9H-PURIN-9-YL]-5-(HYDROXYMETHYL)TETRAHYDRO-3-FURANOL, CITRIC ACID, GLUTAMIC ACID, ... | | Authors: | Moser, J, Schubert, W.-D, Beier, V, Bringemeier, I, Jahn, D, Heinz, D.W. | | Deposit date: | 2001-11-05 | | Release date: | 2002-01-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | V-shaped structure of glutamyl-tRNA reductase, the first enzyme of tRNA-dependent tetrapyrrole biosynthesis.
EMBO J., 20, 2001
|
|
1G9R
 
 | | CRYSTAL STRUCTURE OF GALACTOSYLTRANSFERASE LGTC IN COMPLEX WITH MN AND UDP-2F-GALACTOSE | | Descriptor: | ACETIC ACID, GLYCOSYL TRANSFERASE, MANGANESE (II) ION, ... | | Authors: | Persson, K, Hoa, D.L, Diekelmann, M, Wakarchuk, W.W, Withers, S.G, Strynadka, N.C.J. | | Deposit date: | 2000-11-27 | | Release date: | 2001-02-14 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the retaining galactosyltransferase LgtC from Neisseria meningitidis in complex with donor and acceptor sugar analogs.
Nat.Struct.Biol., 8, 2001
|
|
1GA8
 
 | | CRYSTAL STRUCTURE OF GALACOSYLTRANSFERASE LGTC IN COMPLEX WITH DONOR AND ACCEPTOR SUGAR ANALOGS. | | Descriptor: | 4-deoxy-beta-D-xylo-hexopyranose-(1-4)-beta-D-glucopyranose, GALACTOSYL TRANSFERASE LGTC, MANGANESE (II) ION, ... | | Authors: | Persson, K, Ly, H.D, Diekelmann, M, Wakarchuk, W.W, Withers, S.G, Strynadka, N.C.J. | | Deposit date: | 2000-11-29 | | Release date: | 2001-02-14 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the retaining galactosyltransferase LgtC from Neisseria meningitidis in complex with donor and acceptor sugar analogs.
Nat.Struct.Biol., 8, 2001
|
|
