8B7H
 
 | |
6U9X
 
 | | Structure of T. brucei MERS1-RNA complex | | Descriptor: | Mitochondrial edited mRNA stability factor 1, RNA (5'-R(*GP*AP*GP*AP*GP*GP*GP*GP*GP*UP*U)-3') | | Authors: | Schumacher, M.A. | | Deposit date: | 2019-09-09 | | Release date: | 2019-11-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structures of MERS1, the 5' processing enzyme of mitochondrial mRNAs inTrypanosoma brucei.
Rna, 26, 2020
|
|
4YZV
 
 | | Precleavage 70S structure of the P. vulgaris HigB deltaH92 toxin bound to the ACA codon | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Schureck, M.A, Dunkle, J.A, Maehigashi, T, Dunham, C.M. | | Deposit date: | 2015-03-25 | | Release date: | 2015-10-21 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Defining the mRNA recognition signature of a bacterial toxin protein.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
5HBU
 
 | |
4YPB
 
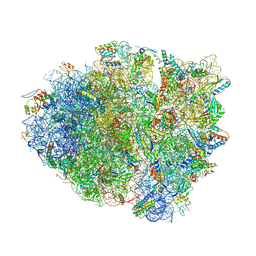 | | Precleavage 70S structure of the P. vulgaris HigB DeltaH92 toxin bound to the AAA codon | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Schureck, M.A, Dunkle, J.A, Maehigashi, T, Dunham, C.M. | | Deposit date: | 2015-03-12 | | Release date: | 2015-10-21 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Defining the mRNA recognition signature of a bacterial toxin protein.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
8TP8
 
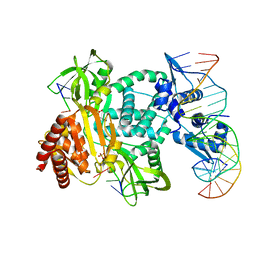 | | Structure of the C. crescentus WYL-activator, DriD, bound to ssDNA and cognate DNA | | Descriptor: | DNA (5'-D(*AP*TP*AP*CP*GP*AP*CP*AP*GP*TP*AP*AP*CP*TP*GP*TP*CP*GP*TP*AP*T)-3'), DNA (5'-D(*AP*TP*AP*CP*GP*AP*CP*AP*GP*TP*TP*AP*CP*TP*GP*TP*CP*GP*TP*AP*T)-3'), DNA (5'-D(P*GP*TP*C)-3'), ... | | Authors: | Schumacher, M.A. | | Deposit date: | 2023-08-04 | | Release date: | 2023-11-29 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Structure of the WYL-domain containing transcription activator, DriD, in complex with ssDNA effector and DNA target site.
Nucleic Acids Res., 52, 2024
|
|
5J6E
 
 | |
5FQL
 
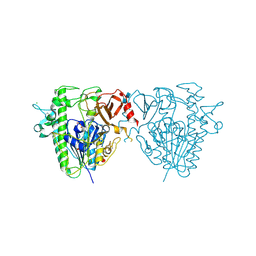 | | Insights into Hunter syndrome from the structure of iduronate-2- sulfatase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Demydchuk, M, Hill, C.H, Zhou, A, Bunkoczi, G, Stein, P.E, Marchesan, D, Deane, J.E, Read, R.J. | | Deposit date: | 2015-12-11 | | Release date: | 2017-01-18 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Insights into Hunter syndrome from the structure of iduronate-2-sulfatase.
Nat Commun, 8, 2017
|
|
7X0F
 
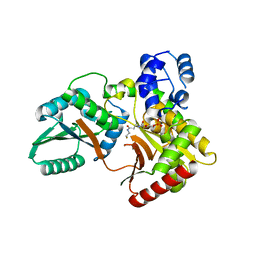 | |
7X0E
 
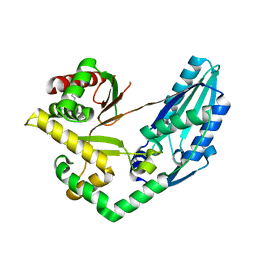 | | Structure of Pseudomonas NRPS protein, AmbB-TC in apo form | | Descriptor: | AMB antimetabolite synthase AmbB, N-methyl-N-[(2S,3R,4R,5R)-2,3,4,5,6-pentakis(oxidanyl)hexyl]nonanamide | | Authors: | ChuYuanKee, M, Bharath, S.R, Song, H. | | Deposit date: | 2022-02-22 | | Release date: | 2022-07-06 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural insights into the substrate-bound condensation domains of non-ribosomal peptide synthetase AmbB.
Sci Rep, 12, 2022
|
|
7X17
 
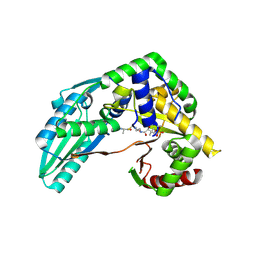 | | Structure of Pseudomonas NRPS protein, AmbB-TC bound to Ppant-L-Ala | | Descriptor: | AMB antimetabolite synthase AmbB, S-[2-[3-[[(2S)-3,3-dimethyl-2-oxidanyl-4-phosphonooxy-butanoyl]amino]propanoylamino]ethyl] (2R)-2-azanylpropanethioate | | Authors: | ChuYuanKee, M, Bharath, S.R, Song, H. | | Deposit date: | 2022-02-23 | | Release date: | 2022-07-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural insights into the substrate-bound condensation domains of non-ribosomal peptide synthetase AmbB.
Sci Rep, 12, 2022
|
|
8TFC
 
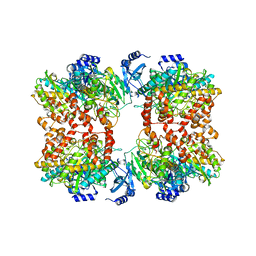 | |
5KGO
 
 | | Structure of K. pneumonia MrkH-c-di-GMP complex | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), Flagellar brake protein YcgR | | Authors: | Schumacher, M. | | Deposit date: | 2016-06-13 | | Release date: | 2016-08-17 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | To be published:
Structures of K. pneumonia MrkH: dual utilization of the PilZ fold for c-di-GMP and DNA binding by a novel activator of biofilm genes
To Be Published
|
|
6CC0
 
 | |
8BAQ
 
 | | E. coli C7 DarT1 in complex with NAD+ | | Descriptor: | 1,2-ETHANEDIOL, DarT ssDNA thymidine ADP-ribosyltransferase family protein, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Schuller, M, Ariza, A. | | Deposit date: | 2022-10-11 | | Release date: | 2023-07-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Molecular basis for the reversible ADP-ribosylation of guanosine bases.
Mol.Cell, 83, 2023
|
|
6CF1
 
 | | Proteus vulgaris HigA antitoxin structure | | Descriptor: | Antitoxin HigA, POTASSIUM ION | | Authors: | Schureck, M.A, Hoffer, E.D, Ei Cho, S, Dunham, C.M. | | Deposit date: | 2018-02-13 | | Release date: | 2019-02-27 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis of transcriptional regulation by the HigA antitoxin.
Mol.Microbiol., 111, 2019
|
|
6UNX
 
 | | Structure of E. coli FtsZ(L178E)-GTP complex | | Descriptor: | Cell division protein FtsZ, GUANOSINE-5'-TRIPHOSPHATE | | Authors: | Schumacher, M.A. | | Deposit date: | 2019-10-13 | | Release date: | 2020-02-05 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | High-resolution crystal structures of Escherichia coli FtsZ bound to GDP and GTP.
Acta Crystallogr.,Sect.F, 76, 2020
|
|
9BE2
 
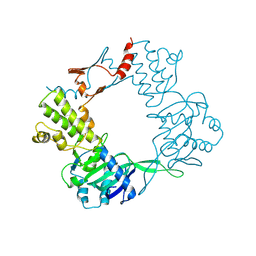 | |
1HY7
 
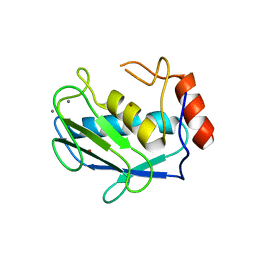 | | A CARBOXYLIC ACID BASED INHIBITOR IN COMPLEX WITH MMP3 | | Descriptor: | CALCIUM ION, R-2-{[4'-METHOXY-(1,1'-BIPHENYL)-4-YL]-SULFONYL}-AMINO-6-METHOXY-HEX-4-YNOIC ACID, STROMELYSIN-1, ... | | Authors: | Natchus, M.G, Bookland, R.G, Laufersweiler, M.J, Pikul, S, Almstead, N.G, De, B, Janusz, M.J, Hsieh, L.C, Gu, F, Pokross, M.E, Patel, V.S, Garver, S.M, Peng, S.X, Branch, T.M, King, S.L, Baker, T.R, Foltz, D.J, Mieling, G.E. | | Deposit date: | 2001-01-18 | | Release date: | 2002-01-18 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Development of new carboxylic acid-based MMP inhibitors derived from functionalized propargylglycines.
J.Med.Chem., 44, 2001
|
|
4YG1
 
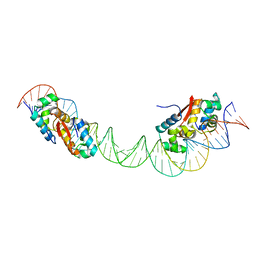 | | HipB-O1-O2 complex/P21212 crystal form | | Descriptor: | Antitoxin HipB, DNA (48-MER) | | Authors: | Schumacher, M.A. | | Deposit date: | 2015-02-25 | | Release date: | 2015-07-29 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | HipBA-promoter structures reveal the basis of heritable multidrug tolerance.
Nature, 524, 2015
|
|
7OMX
 
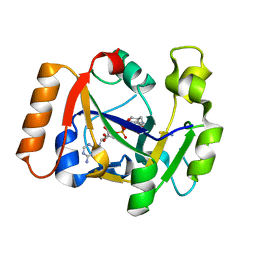 | | Thermus sp. 2.9 DarT in complex with carba-NAD+ | | Descriptor: | CARBA-NICOTINAMIDE-ADENINE-DINUCLEOTIDE, DarT domain-containing protein, THIOCYANATE ION | | Authors: | Schuller, M, Ariza, A. | | Deposit date: | 2021-05-24 | | Release date: | 2021-06-23 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Molecular basis for DarT ADP-ribosylation of a DNA base.
Nature, 596, 2021
|
|
7OMV
 
 | | Thermus sp. 2.9 DarT | | Descriptor: | CHLORIDE ION, DarT domain-containing protein, THIOCYANATE ION | | Authors: | Schuller, M, Ariza, A. | | Deposit date: | 2021-05-24 | | Release date: | 2021-06-23 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | Molecular basis for DarT ADP-ribosylation of a DNA base.
Nature, 596, 2021
|
|
7ON0
 
 | | Thermus sp. 2.9 DarT in complex with ADP-ribosylated ssDNA | | Descriptor: | DNA (5'-D(*AP*TP*GP*TP*C)-3'), DarT domain-containing protein, [(2R,3S,4R,5R)-5-(6-AMINOPURIN-9-YL)-3,4-DIHYDROXY-OXOLAN-2-YL]METHYL [HYDROXY-[[(2R,3S,4R,5S)-3,4,5-TRIHYDROXYOXOLAN-2-YL]METHOXY]PHOSPHORYL] HYDROGEN PHOSPHATE | | Authors: | Schuller, M, Ariza, A. | | Deposit date: | 2021-05-24 | | Release date: | 2021-06-23 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Molecular basis for DarT ADP-ribosylation of a DNA base.
Nature, 596, 2021
|
|
7OMY
 
 | | Thermus sp. 2.9 DarT in complex with carba-NAD+ and ssDNA | | Descriptor: | CARBA-NICOTINAMIDE-ADENINE-DINUCLEOTIDE, DNA (5'-D(*AP*TP*GP*TP*C)-3'), DarT domain-containing protein, ... | | Authors: | Schuller, M, Ariza, A. | | Deposit date: | 2021-05-24 | | Release date: | 2021-06-23 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Molecular basis for DarT ADP-ribosylation of a DNA base.
Nature, 596, 2021
|
|
7OMW
 
 | | Thermus sp. 2.9 DarT in complex with NAD+ | | Descriptor: | DarT domain-containing protein, GLYCEROL, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Schuller, M, Ariza, A. | | Deposit date: | 2021-05-24 | | Release date: | 2021-06-23 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Molecular basis for DarT ADP-ribosylation of a DNA base.
Nature, 596, 2021
|
|
