7TF7
 
 | | S. aureus GS(12) - apo | | Descriptor: | Glutamine synthetase | | Authors: | Travis, B.A, Peck, J, Schumacher, M.A. | | Deposit date: | 2022-01-06 | | Release date: | 2022-06-29 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (2.13 Å) | | Cite: | Molecular dissection of the glutamine synthetase-GlnR nitrogen regulatory circuitry in Gram-positive bacteria.
Nat Commun, 13, 2022
|
|
7TF6
 
 | | S. aureus GS(12)-Q-GlnR peptide | | Descriptor: | GLUTAMINE, Glutamine synthetase, MAGNESIUM ION, ... | | Authors: | Travis, B.A, Peck, J, Schumacher, M.A. | | Deposit date: | 2022-01-06 | | Release date: | 2022-06-29 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (2.15 Å) | | Cite: | Molecular dissection of the glutamine synthetase-GlnR nitrogen regulatory circuitry in Gram-positive bacteria.
Nat Commun, 13, 2022
|
|
7TFE
 
 | | L. monocytogenes GS(12) - apo | | Descriptor: | Glutamine synthetase, MAGNESIUM ION | | Authors: | Travis, B.A, Peck, J, Schumacher, M.A. | | Deposit date: | 2022-01-06 | | Release date: | 2022-06-29 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (2.85 Å) | | Cite: | Molecular dissection of the glutamine synthetase-GlnR nitrogen regulatory circuitry in Gram-positive bacteria.
Nat Commun, 13, 2022
|
|
7TFA
 
 | | P. polymyxa GS(12)-Q-GlnR peptide | | Descriptor: | GLUTAMINE, GlnR C-tail peptide, Glutamine synthetase, ... | | Authors: | Travis, B.A, Peck, J, Schumacher, M.A. | | Deposit date: | 2022-01-06 | | Release date: | 2022-06-29 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (2.07 Å) | | Cite: | Molecular dissection of the glutamine synthetase-GlnR nitrogen regulatory circuitry in Gram-positive bacteria.
Nat Commun, 13, 2022
|
|
7TFC
 
 | | B. subtilis GS(14)-Q-GlnR peptide | | Descriptor: | GLUTAMINE, GlnR C-tail peptide, Glutamine synthetase, ... | | Authors: | Travis, B.A, Peck, J, Schumacher, M.A. | | Deposit date: | 2022-01-06 | | Release date: | 2022-06-29 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (1.96 Å) | | Cite: | Molecular dissection of the glutamine synthetase-GlnR nitrogen regulatory circuitry in Gram-positive bacteria.
Nat Commun, 13, 2022
|
|
7TFD
 
 | | P. polymyxa GS(12) - apo | | Descriptor: | Glutamine synthetase, MAGNESIUM ION | | Authors: | Travis, B.A, Peck, J, Schumacher, M.A. | | Deposit date: | 2022-01-06 | | Release date: | 2022-06-29 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (3.16 Å) | | Cite: | Molecular dissection of the glutamine synthetase-GlnR nitrogen regulatory circuitry in Gram-positive bacteria.
Nat Commun, 13, 2022
|
|
6QWW
 
 | | HEWL lysozyme, crystallized from CuCl2 solution | | Descriptor: | CHLORIDE ION, COPPER (II) ION, Lysozyme C | | Authors: | Boikova, A.S, Dorovatovskii, P.V, Dyakova, Y.A, Ilina, K.B, Kuranova, I.P, Lazarenko, V.A, Marchenkova, M.A, Pisarevsky, Y.V, Timofeev, V.I, Kovalchuk, M.V. | | Deposit date: | 2019-03-06 | | Release date: | 2019-03-13 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | HEWL lysozyme, crystallized from different chlorides
To Be Published
|
|
6QWY
 
 | | HEWL lysozyme, crystallized from NaCl solution | | Descriptor: | CHLORIDE ION, Lysozyme C, SODIUM ION | | Authors: | Boikova, A.S, Dorovatovskii, P.V, Dyakova, Y.A, Ilina, K.B, Kuranova, I.P, Lazarenko, V.A, Marchenkova, M.A, Pisarevsky, Y.V, Timofeev, V.I, Kovalchuk, M.V. | | Deposit date: | 2019-03-06 | | Release date: | 2019-03-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | HEWL lysozyme, crystallized from different chlorides
To Be Published
|
|
6QWZ
 
 | | HEWL lysozyme, crystallized from KCl solution | | Descriptor: | CHLORIDE ION, Lysozyme C | | Authors: | Boikova, A.S, Dorovatovskii, P.V, Dyakova, Y.A, Ilina, K.B, Kuranova, I.P, Lazarenko, V.A, Marchenkova, M.A, Pisarevsky, Y.V, Timofeev, V.I, Kovalchuk, M.V. | | Deposit date: | 2019-03-06 | | Release date: | 2019-03-13 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | HEWL lysozyme, crystallized from different chlorides
To Be Published
|
|
4RMJ
 
 | | Human Sirt2 in complex with ADP ribose and nicotinamide | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, NAD-dependent protein deacetylase sirtuin-2, ... | | Authors: | Rumpf, T, Schiedel, M, Karaman, B, Roessler, C, North, B.J, Lehotzky, A, Olah, J, Ladwein, K.I, Schmidtkunz, K, Gajer, M, Pannek, M, Steegborn, C, Sinclair, D.A, Gerhardt, S, Ovadi, J, Schutkowski, M, Sippl, W, Einsle, O, Jung, M. | | Deposit date: | 2014-10-21 | | Release date: | 2015-02-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Selective Sirt2 inhibition by ligand-induced rearrangement of the active site.
Nat Commun, 6, 2015
|
|
4RMH
 
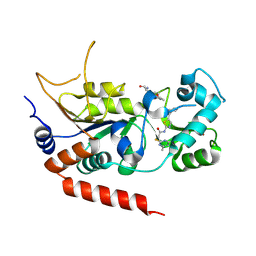 | | Human Sirt2 in complex with SirReal2 and Ac-Lys-H3 peptide | | Descriptor: | 2-[(4,6-dimethylpyrimidin-2-yl)sulfanyl]-N-[5-(naphthalen-1-ylmethyl)-1,3-thiazol-2-yl]acetamide, Ac-Lys-H3 peptide, NAD-dependent protein deacetylase sirtuin-2, ... | | Authors: | Rumpf, T, Schiedel, M, Karaman, B, Roessler, C, North, B.J, Lehotzky, A, Olah, J, Ladwein, K.I, Schmidtkunz, K, Gajer, M, Pannek, M, Steegborn, C, Sinclair, D.A, Gerhardt, S, Ovadi, J, Schutkowski, M, Sippl, W, Einsle, O, Jung, M. | | Deposit date: | 2014-10-21 | | Release date: | 2015-02-25 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Selective Sirt2 inhibition by ligand-induced rearrangement of the active site.
Nat Commun, 6, 2015
|
|
4PYA
 
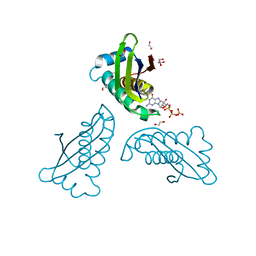 | | MoaC K51A in complex with 3',8-cH2GTP | | Descriptor: | (8S)-3',8-cyclo-7,8-dihydroguanosine 5'-triphosphate, 1,2-ETHANEDIOL, Molybdenum cofactor biosynthesis protein MoaC | | Authors: | Tonthat, N.K, Hover, B.M, Yokoyama, K, Schumacher, M.A. | | Deposit date: | 2014-03-26 | | Release date: | 2015-04-29 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.789 Å) | | Cite: | Mechanism of pyranopterin ring formation in molybdenum cofactor biosynthesis.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4YIY
 
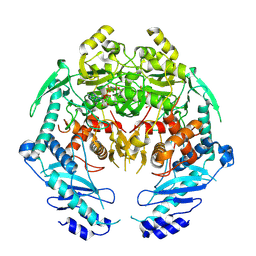 | | Structure of MRB1590 bound to AMP-PNP | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, kRNA Editing A6 Specific Protein | | Authors: | Shaw, P.L.R, Schumacher, M.A. | | Deposit date: | 2015-03-02 | | Release date: | 2015-08-12 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.016 Å) | | Cite: | Structures of the T. brucei kRNA editing factor MRB1590 reveal unique RNA-binding pore motif contained within an ABC-ATPase fold.
Nucleic Acids Res., 43, 2015
|
|
4PYD
 
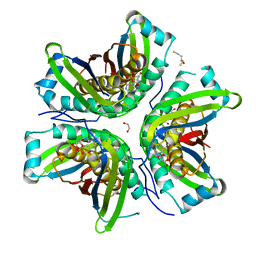 | | MoaC in complex with cPMP crystallized in space group P212121 | | Descriptor: | (2R,4AR,5AR,11AR,12AS)-8-AMINO-2-HYDROXY-4A,5A,9,11,11A,12A-HEXAHYDRO[1,3,2]DIOXAPHOSPHININO[4',5':5,6]PYRANO[3,2-G]PTERIDINE-10,12(4H,6H)-DIONE 2-OXIDE, 1,2-ETHANEDIOL, 2,3-DIHYDROXY-1,4-DITHIOBUTANE, ... | | Authors: | Tonthat, N.K, Hover, B.M, Yokoyama, K, Schumacher, M.A. | | Deposit date: | 2014-03-26 | | Release date: | 2015-04-29 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.186 Å) | | Cite: | Mechanism of pyranopterin ring formation in molybdenum cofactor biosynthesis.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4EO5
 
 | | Yeast Asf1 bound to H3/H4G94P mutant | | Descriptor: | ACETATE ION, GLYCEROL, Histone H3.2, ... | | Authors: | Scorgie, J.K, Churchill, M.E. | | Deposit date: | 2012-04-13 | | Release date: | 2012-06-13 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | The conformational flexibility of the C-terminus of histone H4 promotes histone octamer and nucleosome stability and yeast viability.
Epigenetics Chromatin, 5, 2012
|
|
4ENG
 
 | | STRUCTURE OF ENDOGLUCANASE V CELLOHEXAOSE COMPLEX | | Descriptor: | ENDOGLUCANASE V CELLOHEXAOSE COMPLEX, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Davies, G.J, Schulein, M. | | Deposit date: | 1996-10-17 | | Release date: | 1997-06-16 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure determination and refinement of the Humicola insolens endoglucanase V at 1.5 A resolution.
Acta Crystallogr.,Sect.D, 52, 1996
|
|
5A3H
 
 | | 2-DEOXY-2-FLURO-B-D-CELLOBIOSYL/ENZYME INTERMEDIATE COMPLEX OF THE ENDOGLUCANASE CEL5A FROM BACILLUS AGARADHEARANS AT 1.8 ANGSTROMS RESOLUTION | | Descriptor: | ENDOGLUCANASE, beta-D-glucopyranose-(1-4)-2-deoxy-2-fluoro-alpha-D-glucopyranose | | Authors: | Davies, G.J, Varrot, A, Dauter, M, Brzozowski, A.M, Schulein, M, Mackenzie, L, Withers, S.G. | | Deposit date: | 1998-07-23 | | Release date: | 1999-07-24 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Snapshots along an enzymatic reaction coordinate: analysis of a retaining beta-glycoside hydrolase.
Biochemistry, 37, 1998
|
|
6FTE
 
 | | Crystal structure of an (R)-selective amine transaminase from Exophiala xenobiotica | | Descriptor: | ACETATE ION, Amine transaminase (fold IV), GLYCEROL, ... | | Authors: | Telzerow, A, Hakansson, M, Schurrmann, M, Schwab, H, Steiner, K. | | Deposit date: | 2018-02-21 | | Release date: | 2019-01-09 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Amine Transaminase from Exophiala xenobiotica - Crystal Structure and Engineering of a Fold IV Transaminase that Naturally Converts Biaryl Ketones
Acs Catalysis, 2018
|
|
4YIX
 
 | | Structure of MRB1590 bound to ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, MERCURY (II) ION, ... | | Authors: | Shaw, P.L.R, Schumacher, M.A. | | Deposit date: | 2015-03-02 | | Release date: | 2015-08-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structures of the T. brucei kRNA editing factor MRB1590 reveal unique RNA-binding pore motif contained within an ABC-ATPase fold.
Nucleic Acids Res., 43, 2015
|
|
5HW8
 
 | |
8A3H
 
 | | Cellobiose-derived imidazole complex of the endoglucanase cel5A from Bacillus agaradhaerens at 0.97 A resolution | | Descriptor: | (5R,6R,7R,8S)-7,8-dihydroxy-5-(hydroxymethyl)-5,6,7,8-tetrahydroimidazo[1,2-a]pyridin-6-yl beta-D-glucopyranoside, ACETATE ION, GLYCEROL, ... | | Authors: | Varrot, A, Schulein, M, Pipelier, M, Vasella, A, Davies, G.J. | | Deposit date: | 1999-01-20 | | Release date: | 2000-01-21 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (0.97 Å) | | Cite: | Lateral Protonation of a Glycosidase Inhibitor. Structure of the Bacillus agaradhaerens Cel5A in Complex with a Cellobiose-Derived Imidazole at 0.97 A Resolution
J.Am.Chem.Soc., 121, 1999
|
|
5AHS
 
 | | 3-Sulfinopropionyl-Coenzyme A (3SP-CoA) desulfinase from Advenella mimgardefordensis DPN7T: holo crystal structure with the substrate analog succinyl-CoA | | Descriptor: | ACYL-COA DEHYDROGENASE, COENZYME A, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Cianci, M, Schuermann, M, Meijers, R, Schneider, T.R, Steinbuechel, A. | | Deposit date: | 2015-02-06 | | Release date: | 2015-06-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | 3-Sulfinopropionyl-Coenzyme a (3Sp-Coa) Desulfinase from Advenella Mimigardefordensis Dpn7(T): Crystal Structure and Function of a Desulfinase with an Acyl-Coa Dehydrogenase Fold.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
6HL2
 
 | | wild-type NuoEF from Aquifex aeolicus - oxidized form | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Gerhardt, S, Friedrich, T, Einsle, O, Gnandt, E, Schulte, M, Fiegen, D. | | Deposit date: | 2018-09-10 | | Release date: | 2019-06-26 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | A mechanism to prevent production of reactive oxygen species by Escherichia coli respiratory complex I.
Nat Commun, 10, 2019
|
|
6HL3
 
 | | wild-type NuoEF from Aquifex aeolicus - oxidized form bound to NAD+ | | Descriptor: | 3[N-MORPHOLINO]PROPANE SULFONIC ACID, CHLORIDE ION, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Gerhardt, S, Friedrich, T, Einsle, O, Gnandt, E, Schulte, M, Fiegen, D. | | Deposit date: | 2018-09-10 | | Release date: | 2019-06-26 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | A mechanism to prevent production of reactive oxygen species by Escherichia coli respiratory complex I.
Nat Commun, 10, 2019
|
|
5HW6
 
 | |
