6E71
 
 | | Structure of Human Transthyretin Val30Met/Thr119Met Mutant | | Descriptor: | DI(HYDROXYETHYL)ETHER, Transthyretin | | Authors: | Saelices, L, Chung, K, Sawaya, M.R, Cascio, D, Eisenberg, D. | | Deposit date: | 2018-07-25 | | Release date: | 2019-07-31 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural Variants of Transthyretin
To Be Published
|
|
7RYP
 
 | | Cryo-EM structure of KIFBP:KIF15 | | Descriptor: | KIF-binding protein, Kinesin-like protein KIF15 | | Authors: | Solon, A.L, Tan, Z, Schutt, K.L, Jepsen, L, Haynes, S.E, Nesvizhskii, A.I, Sept, D, Stumpff, J, Ohi, R, Cianfrocco, M.A. | | Deposit date: | 2021-08-25 | | Release date: | 2021-09-08 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Kinesin-binding protein remodels the kinesin motor to prevent microtubule binding.
Sci Adv, 7, 2021
|
|
3J3Y
 
 | |
2WWB
 
 | | CRYO-EM STRUCTURE OF THE MAMMALIAN SEC61 COMPLEX BOUND TO THE ACTIVELY TRANSLATING WHEAT GERM 80S RIBOSOME | | Descriptor: | 25S RRNA, 5.8S RRNA, 60S RIBOSOMAL PROTEIN L17-A, ... | | Authors: | Becker, T, Mandon, E, Bhushan, S, Jarasch, A, Armache, J.P, Funes, S, Jossinet, F, Gumbart, J, Mielke, T, Berninghausen, O, Schulten, K, Westhof, E, Gilmore, R, Beckmann, R. | | Deposit date: | 2009-10-22 | | Release date: | 2009-12-08 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (6.48 Å) | | Cite: | Structure of Monomeric Yeast and Mammalian Sec61 Complexes Interacting with the Translating Ribosome.
Science, 326, 2009
|
|
3UE8
 
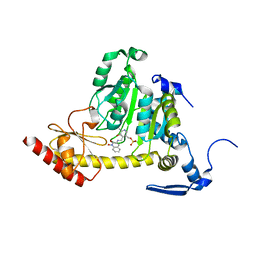 | | Kynurenine Aminotransferase II Inhibitors | | Descriptor: | (5-hydroxy-4-{[(1-hydroxy-2-oxo-1,2-dihydroquinolin-3-yl)amino]methyl}-6-methylpyridin-3-yl)methyl dihydrogen phosphate, CHLORIDE ION, Kynurenine/alpha-aminoadipate aminotransferase, ... | | Authors: | Dounay, A.B, Anderson, M, Bechle, B.M, Campbell, B.M, Claffey, M.M, Evdokimov, A, Edelweiss, E, Fonseca, K.R, Gan, X, Ghosh, S, Hayward, M.M, Horner, W, Kim, J.Y, McAllister, L.A, Pandit, J, Paradis, V, Parikh, V.D, Reese, M.R, Rong, S.B, Salafia, M.A, Schuyten, K, Strick, C.A, Tuttle, J.B, Valentine, J, Wang, H, Zawadzke, L.E, Verhoest, P.R. | | Deposit date: | 2011-10-28 | | Release date: | 2012-02-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.22 Å) | | Cite: | Discovery of Brain-Penetrant, Irreversible Kynurenine Aminotransferase II Inhibitors for Schizophrenia.
ACS Med Chem Lett, 3, 2012
|
|
5EUR
 
 | | Hypothetical protein SF216 from shigella flexneri 5a M90T | | Descriptor: | MAGNESIUM ION, SULFATE ION, Uncharacterized protein | | Authors: | Lee, Y.-S, Seok, S.-H, Kim, H.-N, An, J.-G, Chung, K.Y, Won, H.-S, Seo, M.-D. | | Deposit date: | 2015-11-19 | | Release date: | 2016-11-23 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.698 Å) | | Cite: | Crystal structure of SF216 from Shigella flexneri strain 5a
To Be Published
|
|
2WW9
 
 | | Cryo-EM structure of the active yeast Ssh1 complex bound to the yeast 80S ribosome | | Descriptor: | 25S RRNA, 60S RIBOSOMAL PROTEIN L17-A, 60S RIBOSOMAL PROTEIN L19, ... | | Authors: | Becker, T, Mandon, E, Bhushan, S, Jarasch, A, Armache, J.P, Funes, S, Jossinet, F, Gumbart, J, Mielke, T, Berninghausen, O, Schulten, K, Westhof, E, Gilmore, R, Beckmann, R. | | Deposit date: | 2009-10-22 | | Release date: | 2009-12-08 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (8.6 Å) | | Cite: | Structure of Monomeric Yeast and Mammalian Sec61 Complexes Interacting with the Translating Ribosome.
Science, 326, 2009
|
|
2XBM
 
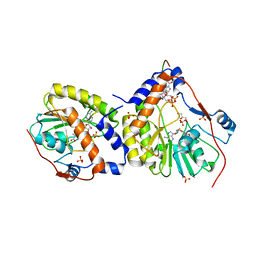 | | Crystal structure of the dengue virus methyltransferase bound to a 5'- capped octameric RNA | | Descriptor: | 5'-(*G3AP*GP*AP*AP*CP*CP*UP*GP*A)-3', GLYCEROL, NONSTRUCTURAL PROTEIN NS5, ... | | Authors: | Yap, L.J, Luo, D.H, Chung, K.Y, Lim, S.P, Bodenreider, C, Noble, C, Shi, P.Y, Lescar, J. | | Deposit date: | 2010-04-13 | | Release date: | 2010-09-29 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal Structure of the Dengue Virus Methyltransferase Bound to a 5'-Capped Octameric RNA
Plos One, 5, 2010
|
|
5FFR
 
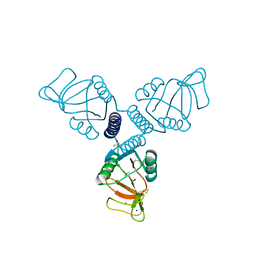 | | Crystal Structure of Surfactant Protein-A complexed with phosphocholine | | Descriptor: | CALCIUM ION, PHOSPHOCHOLINE, Pulmonary surfactant-associated protein A, ... | | Authors: | Goh, B.C, Wu, H, Rynkiewicz, M.J, Schulten, K, Seaton, B.A, McCormack, F.X. | | Deposit date: | 2015-12-18 | | Release date: | 2016-07-06 | | Last modified: | 2019-12-11 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Elucidation of Lipid Binding Sites on Lung Surfactant Protein A Using X-ray Crystallography, Mutagenesis, and Molecular Dynamics Simulations.
Biochemistry, 55, 2016
|
|
5FJB
 
 | | Cyclophilin A Stabilize HIV-1 Capsid through a Novel Non- canonical Binding Site | | Descriptor: | GAG POLYPROTEIN, PEPTIDYL-PROLYL CIS-TRANS ISOMERASE A | | Authors: | Liu, C, Perilla, J.R, Ning, J, Lu, M, Hou, G, Ramalhu, R, Bedwell, G.J, Ahn, J, Shi, J, Gronenborn, A.M, Prevelige Jr, P.E, Rousso, I, Aiken, C, Polenova, T, Schulten, K, Zhang, P. | | Deposit date: | 2015-10-07 | | Release date: | 2016-03-16 | | Last modified: | 2017-08-23 | | Method: | ELECTRON MICROSCOPY (9 Å) | | Cite: | Cyclophilin a Stabilizes the HIV-1 Capsid Through a Novel Non-Canonical Binding Site.
Nat.Commun., 7, 2016
|
|
2JMH
 
 | | NMR solution structure of Blo t 5, a major mite allergen from Blomia tropicalis | | Descriptor: | Mite allergen Blo t 5 | | Authors: | Naik, M.T, Chang, C, Kuo, I, Chua, K, Huang, T. | | Deposit date: | 2006-11-12 | | Release date: | 2007-11-13 | | Last modified: | 2023-12-20 | | Method: | SOLUTION NMR | | Cite: | Roles of Structure and Structural Dynamics in the Antibody Recognition of the Allergen Proteins: An NMR Study on Blomia tropicalis Major Allergen
Structure, 16, 2008
|
|
5FFS
 
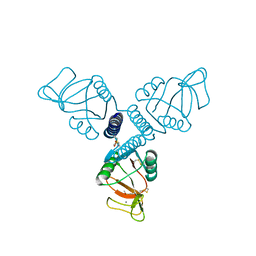 | | Crystal Structure of Surfactant Protein-A Y164A Mutant | | Descriptor: | CALCIUM ION, Pulmonary surfactant-associated protein A | | Authors: | Goh, B.C, Wu, H, Rynkiewicz, M.J, Schulten, K, Seaton, B.A, McCormack, F.X. | | Deposit date: | 2015-12-18 | | Release date: | 2016-07-06 | | Last modified: | 2019-12-11 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Elucidation of Lipid Binding Sites on Lung Surfactant Protein A Using X-ray Crystallography, Mutagenesis, and Molecular Dynamics Simulations.
Biochemistry, 55, 2016
|
|
5FFT
 
 | | Crystal Structure of Surfactant Protein-A Y221A Mutant | | Descriptor: | CALCIUM ION, Pulmonary surfactant-associated protein A | | Authors: | Goh, B.C, Wu, H, Rynkiewicz, M.J, Schulten, K, Seaton, B.A, McCormack, F.X. | | Deposit date: | 2015-12-18 | | Release date: | 2016-07-06 | | Last modified: | 2019-12-11 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Elucidation of Lipid Binding Sites on Lung Surfactant Protein A Using X-ray Crystallography, Mutagenesis, and Molecular Dynamics Simulations.
Biochemistry, 55, 2016
|
|
2WWA
 
 | | Cryo-EM structure of idle yeast Ssh1 complex bound to the yeast 80S ribosome | | Descriptor: | 25S RRNA, 60S RIBOSOMAL PROTEIN L17-A, 60S RIBOSOMAL PROTEIN L19, ... | | Authors: | Becker, T, Mandon, E, Bhushan, S, Jarasch, A, Armache, J.P, Funes, S, Jossinet, F, Gumbart, J, Mielke, T, Berninghausen, O, Schulten, K, Westhof, E, Gilmore, R, Beckmann, R. | | Deposit date: | 2009-10-22 | | Release date: | 2009-12-08 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (8.9 Å) | | Cite: | Structure of Monomeric Yeast and Mammalian Sec61 Complexes Interacting with the Translating Ribosome.
Science, 326, 2009
|
|
3G46
 
 | | Ligand migration and cavities within scapharca dimeric hemoglobin: wild type with co bound to heme and chloroform bound to the XE4 cavity | | Descriptor: | CARBON MONOXIDE, Globin-1, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Knapp, J.E, Pahl, R, Cohen, J, Nichols, J.C, Schulten, K, Gibson, Q.H, Srajer, V, Royer Jr, W.E. | | Deposit date: | 2009-02-03 | | Release date: | 2009-12-01 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (0.91 Å) | | Cite: | Ligand migration and cavities within Scapharca Dimeric HbI: studies by time-resolved crystallo-graphy, Xe binding, and computational analysis.
Structure, 17, 2009
|
|
3G4Q
 
 | | Ligand migration and cavities within scapharca dimeric hemoglobin: wild type with co bound to heme and chloroform bound to the XE4 cavity | | Descriptor: | CARBON MONOXIDE, Globin-1, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Knapp, J.E, Pahl, R, Cohen, J, Nichols, J.C, Schulten, K, Gibson, Q.H, Srajer, V, Royer Jr, W.E. | | Deposit date: | 2009-02-04 | | Release date: | 2009-12-01 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Ligand migration and cavities within Scapharca Dimeric HbI: studies by time-resolved crystallo-graphy, Xe binding, and computational analysis.
Structure, 17, 2009
|
|
1ODS
 
 | | Cephalosporin C deacetylase from Bacillus subtilis | | Descriptor: | CEPHALOSPORIN C DEACETYLASE, CHLORIDE ION, MAGNESIUM ION | | Authors: | Vincent, F, Charnock, S.J, Verschueren, K.H.G, Turkenburg, J.P, Scott, D.J, Offen, W.A, Roberts, S, Pell, G, Gilbert, H.J, Brannigan, J.A, Davies, G.J. | | Deposit date: | 2003-02-20 | | Release date: | 2003-07-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Multifunctional Xylooligosaccharide/Cephalosporin C Deacetylase Revealed by the Hexameric Structure of the Bacillus Subtilis Enzyme at 1.9A Resolution
J.Mol.Biol., 330, 2003
|
|
3J4F
 
 | | Structure of HIV-1 capsid protein by cryo-EM | | Descriptor: | capsid protein | | Authors: | Zhao, G, Perilla, J.R, Meng, X, Schulten, K, Zhang, P. | | Deposit date: | 2013-07-11 | | Release date: | 2013-07-24 | | Last modified: | 2018-07-18 | | Method: | ELECTRON MICROSCOPY (8.6 Å) | | Cite: | Mature HIV-1 capsid structure by cryo-electron microscopy and all-atom molecular dynamics.
Nature, 497, 2013
|
|
1OSY
 
 | | Crystal structure of FIP-Fve fungal immunomodulatory protein | | Descriptor: | BROMIDE ION, IMMUNOMODULATORY PROTEIN FIP-FVE | | Authors: | Palasingam, P, Joseph, J.S, Seow, S.V, Shai, V, Robinson, H, Chua, K.Y, Kolatkar, P.R. | | Deposit date: | 2003-03-20 | | Release date: | 2003-11-04 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A 1.7A structure of Fve, a member of the new fungal
immunomodulatory protein family
J.Mol.Biol., 332, 2003
|
|
2X6M
 
 | | Structure of a single domain camelid antibody fragment in complex with a C-terminal peptide of alpha-synuclein | | Descriptor: | ALPHA-SYNUCLEIN PEPTIDE, HEAVY CHAIN VARIABLE DOMAIN FROM DROMEDARY | | Authors: | DeGenst, E, Guilliams, T, Wellens, J, O'Day, E.M, Waudby, C.A, Meehan, S, Dumoulin, M, Hsu, S.-T.D, Cremades, N, Verschueren, K.H.G, Pardon, E, Wyns, L, Steyaert, J, Christodoulou, J, Dobson, C.M. | | Deposit date: | 2010-02-18 | | Release date: | 2010-06-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Structure and Properties of a Complex of Alpha-Synuclein and a Single-Domain Camelid Antibody.
J.Mol.Biol., 402, 2010
|
|
1OWW
 
 | | Solution structure of the first type III module of human fibronectin determined by 1H, 15N NMR spectroscopy | | Descriptor: | Fibronectin first type III module | | Authors: | Gao, M, Craig, D, Lequin, O, Campbell, I.D, Vogel, V, Schulten, K. | | Deposit date: | 2003-03-31 | | Release date: | 2003-10-07 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure and functional significance of mechanically unfolded fibronectin type III1 intermediates
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
3G4Y
 
 | | Ligand migration and cavities within scapharca dimeric hemoglobin: wild type with co bound to heme and chloromethyl benzene bound to the XE4 cavity | | Descriptor: | (chloromethyl)benzene, CARBON MONOXIDE, GLOBIN-1, ... | | Authors: | Knapp, J.E, Pahl, R, Cohen, J, Nichols, J.C, Schulten, K, Gibson, Q.H, Srajer, V, Royer Jr, W.E. | | Deposit date: | 2009-02-04 | | Release date: | 2009-12-01 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Ligand migration and cavities within Scapharca Dimeric HbI: studies by time-resolved crystallo-graphy, Xe binding, and computational analysis.
Structure, 17, 2009
|
|
3G53
 
 | | Ligand migration and cavities within scapharca dimeric hemoglobin: wild type with co bound to heme and chloropropyl benzene bound to the XE4 cavity | | Descriptor: | (3-chloropropyl)benzene, CARBON MONOXIDE, Globin-1, ... | | Authors: | Knapp, J.E, Pahl, R, Cohen, J, Nichols, J.C, Schulten, K, Gibson, Q.H, Srajer, V, Royer Jr, W.E. | | Deposit date: | 2009-02-04 | | Release date: | 2009-12-01 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Ligand migration and cavities within Scapharca Dimeric HbI: studies by time-resolved crystallo-graphy, Xe binding, and computational analysis.
Structure, 17, 2009
|
|
3G4U
 
 | | Ligand migration and cavities within scapharca dimeric hemoglobin: wild type with co bound to heme and dichloropropane bound to the XE4 cavity | | Descriptor: | 1,3-dichloropropane, CARBON MONOXIDE, GLOBIN-1, ... | | Authors: | Knapp, J.E, Pahl, R, Cohen, J, Nichols, J.C, Schulten, K, Gibson, Q.H, Srajer, V, Royer Jr, W.E. | | Deposit date: | 2009-02-04 | | Release date: | 2009-12-01 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Ligand migration and cavities within Scapharca Dimeric HbI: studies by time-resolved crystallo-graphy, Xe binding, and computational analysis.
Structure, 17, 2009
|
|
3G4V
 
 | | Ligand migration and cavities within scapharca dimeric hemoglobin: wild type with co bound to heme and chloropentane bound to the XE4 cavity | | Descriptor: | 1-chloropentane, CARBON MONOXIDE, GLOBIN-1, ... | | Authors: | Knapp, J.E, Pahl, R, Cohen, J, Nichols, J.C, Schulten, K, Gibson, Q.H, Srajer, V, Royer Jr, W.E. | | Deposit date: | 2009-02-04 | | Release date: | 2009-12-01 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Ligand migration and cavities within Scapharca Dimeric HbI: studies by time-resolved crystallo-graphy, Xe binding, and computational analysis.
Structure, 17, 2009
|
|
