2NMX
 
 | | Structure of inhibitor binding to Carbonic Anhydrase I | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Carbonic anhydrase 1, N-{2-[4-(AMINOSULFONYL)PHENYL]ETHYL}ACETAMIDE, ... | | Authors: | Christianson, D.W, Jude, K.M. | | Deposit date: | 2006-10-23 | | Release date: | 2007-04-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural Analysis of Charge Discrimination in the Binding of Inhibitors to Human Carbonic Anhydrases I and II
J.Am.Chem.Soc., 129, 2007
|
|
2NN7
 
 | | Structure of inhibitor binding to Carbonic Anhydrase I | | Descriptor: | Carbonic anhydrase 1, DIMETHYL SULFOXIDE, ETHYL 3-[4-(AMINOSULFONYL)PHENYL]PROPANOATE, ... | | Authors: | Christianson, D.W, Jude, K.M. | | Deposit date: | 2006-10-23 | | Release date: | 2007-05-08 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural Analysis of Charge Discrimination in the Binding of Inhibitors to Human Carbonic Anhydrases I and II.
J.Am.Chem.Soc., 129, 2007
|
|
2NNG
 
 | | Structure of inhibitor binding to Carbonic Anhydrase II | | Descriptor: | 4-(2-AMINOETHYL)BENZENESULFONAMIDE, Carbonic anhydrase 2, GLYCEROL, ... | | Authors: | Christianson, D.W, Jude, K.M. | | Deposit date: | 2006-10-24 | | Release date: | 2007-05-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structural Analysis of Charge Discrimination in the Binding of Inhibitors to Human Carbonic Anhydrases I and II.
J.Am.Chem.Soc., 129, 2007
|
|
2NNS
 
 | |
2NN1
 
 | | Structure of inhibitor binding to Carbonic Anhydrase I | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3-[4-(AMINOSULFONYL)PHENYL]PROPANOIC ACID, Carbonic anhydrase 1, ... | | Authors: | Christianson, D.W, Jude, K.M. | | Deposit date: | 2006-10-23 | | Release date: | 2007-04-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural Analysis of Charge Discrimination in the Binding of Inhibitors to Human Carbonic Anhydrases I and II
J.Am.Chem.Soc., 129, 2007
|
|
2NNO
 
 | | Structure of inhibitor binding to Carbonic Anhydrase II | | Descriptor: | 3-[4-(AMINOSULFONYL)PHENYL]PROPANOIC ACID, Carbonic anhydrase 2, GLYCEROL, ... | | Authors: | Christianson, D.W, Jude, K.M. | | Deposit date: | 2006-10-24 | | Release date: | 2007-05-08 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.01 Å) | | Cite: | Structural Analysis of Charge Discrimination in the Binding of Inhibitors to Human Carbonic Anhydrases I and II.
J.Am.Chem.Soc., 129, 2007
|
|
2NNV
 
 | | Structure of inhibitor binding to Carbonic Anhydrase II | | Descriptor: | Carbonic anhydrase 2, ETHYL 3-[4-(AMINOSULFONYL)PHENYL]PROPANOATE, GLYCEROL, ... | | Authors: | Christianson, D.W, Jude, K.M. | | Deposit date: | 2006-10-24 | | Release date: | 2007-05-08 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Structural Analysis of Charge Discrimination in the Binding of Inhibitors to Human Carbonic Anhydrases I and II.
J.Am.Chem.Soc., 129, 2007
|
|
6V0K
 
 | |
8V3K
 
 | | F95S-F198S epi-Isozizaene Synthase: complex with 3 Mg2+, inorganic pyrophosphate, and benzyl triethyl ammonium cation | | Descriptor: | DIPHOSPHATE, Epi-isozizaene synthase, GLYCEROL, ... | | Authors: | Christianson, D.W, Eaton, S.A. | | Deposit date: | 2023-11-28 | | Release date: | 2024-01-24 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Structure-Based Engineering of a Sesquiterpene Cyclase to Generate an Alcohol Product: Conversion of epi -Isozizaene Synthase into alpha-Bisabolol Synthase.
Biochemistry, 63, 2024
|
|
5VI6
 
 | | Crystal structure of histone deacetylase 8 in complex with trapoxin A | | Descriptor: | 1,2-ETHANEDIOL, 1,4-DIETHYLENE DIOXIDE, Histone deacetylase 8, ... | | Authors: | Porter, N.J, Christianson, D.W. | | Deposit date: | 2017-04-14 | | Release date: | 2017-09-06 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.237 Å) | | Cite: | Binding of the Microbial Cyclic Tetrapeptide Trapoxin A to the Class I Histone Deacetylase HDAC8.
ACS Chem. Biol., 12, 2017
|
|
5HJA
 
 | | Crystal structure of Leishmania mexicana arginase in complex with inhibitor ABHDP | | Descriptor: | (R)-2-amino-6-borono-2-(1-(3,4-dichlorobenzyl)piperidin-4-yl)hexanoic acid, Arginase, GLYCEROL, ... | | Authors: | Hai, Y, Christianson, D.W. | | Deposit date: | 2016-01-13 | | Release date: | 2016-04-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structures of Leishmania mexicana arginase complexed with alpha , alpha-disubstituted boronic amino-acid inhibitors.
Acta Crystallogr F Struct Biol Commun, 72, 2016
|
|
3CAA
 
 | | CLEAVED ANTICHYMOTRYPSIN A347R | | Descriptor: | ANTICHYMOTRYPSIN | | Authors: | Lukacs, C.M, Christianson, D.W. | | Deposit date: | 1997-08-18 | | Release date: | 1998-02-25 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Engineering an anion-binding cavity in antichymotrypsin modulates the "spring-loaded" serpin-protease interaction.
Biochemistry, 37, 1998
|
|
1VJ5
 
 | | Human soluble Epoxide Hydrolase- N-cyclohexyl-N'-(4-iodophenyl)urea complex | | Descriptor: | HEXAETHYLENE GLYCOL, MAGNESIUM ION, N-CYCLOHEXYL-N'-(4-IODOPHENYL)UREA, ... | | Authors: | Gomez, G.A, Morisseau, C, Hammock, B.D, Christianson, D.W. | | Deposit date: | 2004-02-03 | | Release date: | 2004-04-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structure of human epoxide hydrolase reveals mechanistic inferences on bifunctional catalysis in epoxide and phosphate ester hydrolysis
Biochemistry, 43, 2004
|
|
7CA2
 
 | |
3CYU
 
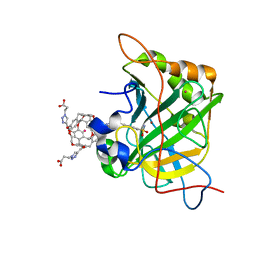 | | Human Carbonic Anhydrase II complexed with Cryptophane biosensor and xenon | | Descriptor: | Carbonic anhydrase 2, MoMo-2-[4-(2-(4-(methoxy)-1H-1,2,3-triazol-1-yl)ethyl)benzenesulfonamide]-7,12-bis-[3-(4-(methoxy)-1H-1,2,3-triazol-1-y l)propanoic acid]-cryptophane-A, PoPo-2-[4-(2-(4-(methoxy)-1H-1,2,3-triazol-1-yl)ethyl)benzenesulfonamide]-7,12-bis-[3-(4-(methoxy)-1H-1,2,3-triazol-1-yl)propanoic acid]-cryptophane-A, ... | | Authors: | Aaron, J.A, Jude, K.M, Di Costanzo, L, Christianson, D.W. | | Deposit date: | 2008-04-26 | | Release date: | 2008-05-27 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of a 129Xe-cryptophane biosensor complexed with human carbonic anhydrase II.
J.Am.Chem.Soc., 130, 2008
|
|
6DVM
 
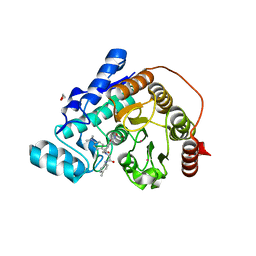 | | Crystal structure of Danio rerio histone deacetylase 6 catalytic domain 2 in complex with DDK-122 | | Descriptor: | 1,2-ETHANEDIOL, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 4-(dimethylamino)-N-{[4-(hydroxycarbamoyl)phenyl]methyl}-N-{2-[(4-methylphenyl)amino]-2-oxoethyl}benzamide, ... | | Authors: | Porter, N.J, Christianson, D.W. | | Deposit date: | 2018-06-24 | | Release date: | 2018-08-29 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Histone Deacetylase 6-Selective Inhibitors and the Influence of Capping Groups on Hydroxamate-Zinc Denticity.
J. Med. Chem., 61, 2018
|
|
6DVO
 
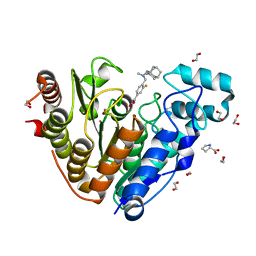 | | Crystal structure of Danio rerio histone deacetylase 6 catalytic domain 2 in complex with Bavarostat | | Descriptor: | 1,2-ETHANEDIOL, 3-fluoro-N-hydroxy-4-[(methyl{[(3s,5s,7s)-tricyclo[3.3.1.1~3,7~]decan-1-yl]methyl}amino)methyl]benzamide, Hdac6 protein, ... | | Authors: | Porter, N.J, Christianson, D.W. | | Deposit date: | 2018-06-24 | | Release date: | 2018-08-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Histone Deacetylase 6-Selective Inhibitors and the Influence of Capping Groups on Hydroxamate-Zinc Denticity.
J. Med. Chem., 61, 2018
|
|
9CA2
 
 | |
3V1V
 
 | |
3V1X
 
 | |
5D1B
 
 | | Crystal structure of G117E HDAC8 in complex with TSA | | Descriptor: | Histone deacetylase 8, POTASSIUM ION, TRICHOSTATIN A, ... | | Authors: | Decroos, C, Christianson, N.H, Gullett, L.E, Bowman, C.M, Christianson, K.E, Deardorff, M.A, Christianson, D.W. | | Deposit date: | 2015-08-04 | | Release date: | 2015-10-21 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Biochemical and Structural Characterization of HDAC8 Mutants Associated with Cornelia de Lange Syndrome Spectrum Disorders.
Biochemistry, 54, 2015
|
|
5D1D
 
 | | Crystal structure of P91L-Y306F HDAC8 in complex with a tetrapeptide substrate | | Descriptor: | HDAC8 Fluor de Lys tetrapeptide substrate, Histone deacetylase 8, POTASSIUM ION, ... | | Authors: | Decroos, C, Christianson, N.H, Gullett, L.E, Bowman, C.M, Christianson, K.E, Deardorff, M.A, Christianson, D.W. | | Deposit date: | 2015-08-04 | | Release date: | 2015-10-21 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.011 Å) | | Cite: | Biochemical and Structural Characterization of HDAC8 Mutants Associated with Cornelia de Lange Syndrome Spectrum Disorders.
Biochemistry, 54, 2015
|
|
7S0L
 
 | |
7S0H
 
 | |
7S0A
 
 | |
