1L1V
 
 | | UNUSUAL ACTD/DNA_TA COMPLEX STRUCTURE | | Descriptor: | 5'-D(*GP*TP*CP*AP*CP*CP*GP*AP*C)-3', ACTINOMYCIN D | | Authors: | Chou, S.-H, Chin, K.-H, Chen, F.-M. | | Deposit date: | 2002-02-20 | | Release date: | 2002-03-06 | | Last modified: | 2024-07-10 | | Method: | SOLUTION NMR | | Cite: | Looped Out and Perpendicular: Deformation of Watson-Crick Base Pair Associated with Actinomycin D Binding.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
8E9B
 
 | |
1D69
 
 | |
6DJO
 
 | | Cryo-EM structure of ADP-actin filaments | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Chou, S.Z, Pollard, T.D. | | Deposit date: | 2018-05-25 | | Release date: | 2019-02-27 | | Last modified: | 2024-04-10 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Mechanism of actin polymerization revealed by cryo-EM structures of actin filaments with three different bound nucleotides.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6DJM
 
 | | Cryo-EM structure of AMPPNP-actin filaments | | Descriptor: | Actin, alpha skeletal muscle, MAGNESIUM ION, ... | | Authors: | Chou, S.Z, Pollard, T.D. | | Deposit date: | 2018-05-25 | | Release date: | 2019-02-27 | | Last modified: | 2020-01-08 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Mechanism of actin polymerization revealed by cryo-EM structures of actin filaments with three different bound nucleotides.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6DJN
 
 | | Cryo-EM structure of ADP-Pi-actin filaments | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Chou, S.Z, Pollard, T.D. | | Deposit date: | 2018-05-25 | | Release date: | 2019-02-27 | | Last modified: | 2020-01-08 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Mechanism of actin polymerization revealed by cryo-EM structures of actin filaments with three different bound nucleotides.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
103D
 
 | |
1BJH
 
 | | HAIRPIN LOOPS CONSISTING OF SINGLE ADENINE RESIDUES CLOSED BY SHEARED A(DOT)A AND G(DOT)G PAIRS FORMED BY THE DNA TRIPLETS AAA AND GAG: SOLUTION STRUCTURE OF THE D(GTACAAAGTAC) HAIRPIN, NMR, 16 STRUCTURES | | Descriptor: | DNA (5'-D(*GP*TP*AP*CP*AP*AP*AP*GP*TP*AP*C)-3') | | Authors: | Chou, S.-H, Zhu, L, Gao, Z, Cheng, J.-W, Reid, B.R. | | Deposit date: | 1997-07-25 | | Release date: | 1997-12-03 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Hairpin loops consisting of single adenine residues closed by sheared A.A and G.G pairs formed by the DNA triplets AAA and GAG: solution structure of the d(GTACAAAGTAC) hairpin.
J.Mol.Biol., 264, 1996
|
|
175D
 
 | |
4EOB
 
 | |
7K20
 
 | |
4F4M
 
 | |
7K21
 
 | |
3MHX
 
 | | Crystal Structure of Stenotrophomonas maltophilia FeoA complexed with Zinc: A Unique Procaryotic SH3 Domain Protein Possibly Acting as a Bacterial Ferrous Iron Transport Activating Factor | | Descriptor: | ACETATE ION, CHLORIDE ION, Putative ferrous iron transport protein A, ... | | Authors: | Chou, S.-H, Su, Y.-C, Chin, K.-H, Hung, H.-C, Shen, G.-H, Wang, A.H.-J. | | Deposit date: | 2010-04-09 | | Release date: | 2010-06-23 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of Stenotrophomonas maltophilia FeoA complexed with zinc: a unique prokaryotic SH3-domain protein that possibly acts as a bacterial ferrous iron-transport activating factor
Acta Crystallogr.,Sect.F, 66, 2010
|
|
3RMR
 
 | |
3M4U
 
 | |
6WIN
 
 | | Type 6 secretion amidase effector 2 (Tae2) | | Descriptor: | Type 6 secretion amidase effector 2 | | Authors: | Chou, S, Radkov, A.D. | | Deposit date: | 2020-04-10 | | Release date: | 2020-04-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Ticks Resist Skin Commensals with Immune Factor of Bacterial Origin.
Cell, 183, 2020
|
|
3C12
 
 | |
2FA5
 
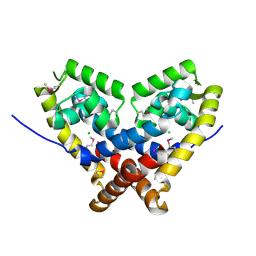 | | The crystal structure of an unliganded multiple antibiotic-resistance repressor (MarR) from Xanthomonas campestris | | Descriptor: | CHLORIDE ION, transcriptional regulator marR/emrR family | | Authors: | Chin, K.H, Tu, Z.L, Li, J.N, Chou, C.C, Wang, A.H.J, Chou, S.H. | | Deposit date: | 2005-12-06 | | Release date: | 2006-11-14 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The crystal structure of XC1739: a putative multiple antibiotic-resistance repressor (MarR) from Xanthomonas campestris at 1.8 A resolution
Proteins, 65, 2006
|
|
2FUJ
 
 | |
2GU9
 
 | |
2GBZ
 
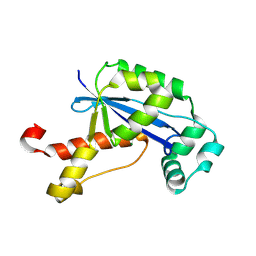 | | The Crystal Structure of XC847 from Xanthomonas campestris: a 3-5 Oligoribonuclease of DnaQ fold family with a Novel Opposingly-Shifted Helix | | Descriptor: | MAGNESIUM ION, Oligoribonuclease | | Authors: | Chin, K.H, Yang, C.Y, Chou, C.C, Wang, A.H.J, Chou, S.H. | | Deposit date: | 2006-03-12 | | Release date: | 2007-01-16 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The crystal structure of XC847 from Xanthomonas campestris: a 3'-5' oligoribonuclease of DnaQ fold family with a novel opposingly shifted helix
Proteins, 65, 2006
|
|
2FUK
 
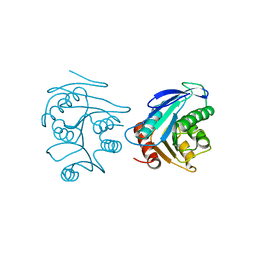 | | Crystal structure of XC6422 from Xanthomonas campestris: a member of a/b serine hydrolase without lid at 1.6 resolution | | Descriptor: | XC6422 protein | | Authors: | Yang, C.Y, Chin, K.H, Chou, C.C, Wang, A.H.J, Chou, S.H. | | Deposit date: | 2006-01-27 | | Release date: | 2006-07-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of XC6422 from Xanthomonas campestris at 1.6 A resolution: a small serine alpha/beta-hydrolase
Acta Crystallogr.,Sect.F, 62, 2006
|
|
1OVF
 
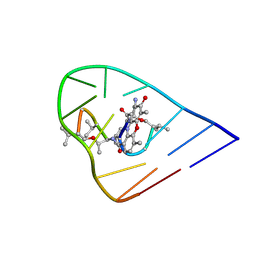 | | NMR Structure of ActD/5'-CCGTTTTGTGG-3' Complex | | Descriptor: | (5'-D(*CP*CP*GP*TP*TP*TP*TP*GP*TP*GP*G)-3'), ACTINOMYCIN D | | Authors: | Chin, K.-H, Chou, S.-H, Chen, F.-M. | | Deposit date: | 2003-03-26 | | Release date: | 2003-05-27 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the Actd-5'-Ccgtt(3)Gtgg-3' Complex: Drug Interaction with Tandem G.T Mismatches and Hairpin Loop Backbone.
Nucleic Acids Res., 31, 2003
|
|
1P0U
 
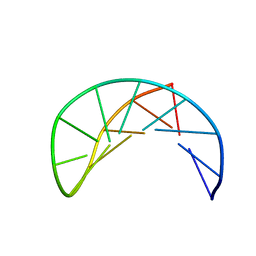 | | Sheared G/C Base Pair | | Descriptor: | 5'-D(*GP*CP*AP*TP*CP*GP*AP*CP*GP*AP*TP*GP*C)-3' | | Authors: | Chin, K.H, Chou, S.H. | | Deposit date: | 2003-04-11 | | Release date: | 2003-06-03 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Sheared-type G(anti).C(syn) Base-pair: A Unique d(GXC) Loop Closure Motif.
J.Mol.Biol., 329, 2003
|
|
