4OYH
 
 | | Structure of Bacillus subtilis MobB | | Descriptor: | Molybdopterin-guanine dinucleotide biosynthesis protein B, SULFATE ION | | Authors: | Kim, D, Choe, J. | | Deposit date: | 2014-02-12 | | Release date: | 2015-03-18 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.409 Å) | | Cite: | Bacillus subtilis MobB
To Be Published
|
|
5U6N
 
 | | Crystal structure of UDP-glucosyltransferase, UGT74F2 (T15S), with UDP and salicylic acid | | Descriptor: | 2-HYDROXYBENZOIC ACID, GLUTAMINE, UDP-glycosyltransferase 74F2, ... | | Authors: | George Thompson, A.M, Iancu, C.V, Dean, J.V, Choe, J. | | Deposit date: | 2016-12-08 | | Release date: | 2017-05-03 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Differences in salicylic acid glucose conjugations by UGT74F1 and UGT74F2 from Arabidopsis thaliana.
Sci Rep, 7, 2017
|
|
5V2J
 
 | | Crystal structure of UDP-glucosyltransferase, UGT74F2 (T15S), with UDP and 2-bromobenzoic acid | | Descriptor: | 2-bromobenzoic acid, UDP-glycosyltransferase 74F2, URIDINE-5'-DIPHOSPHATE, ... | | Authors: | George Thompson, A.M, Iancu, C.V, Dean, J.V, Choe, J. | | Deposit date: | 2017-03-04 | | Release date: | 2017-05-03 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Differences in salicylic acid glucose conjugations by UGT74F1 and UGT74F2 from Arabidopsis thaliana.
Sci Rep, 7, 2017
|
|
5V2K
 
 | | Crystal structure of UDP-glucosyltransferase, UGT74F2 (T15A), with UDP and 2-bromobenzoic acid | | Descriptor: | 2-bromobenzoic acid, UDP-glycosyltransferase 74F2, URIDINE-5'-DIPHOSPHATE, ... | | Authors: | George Thompson, A.M, Iancu, C.V, Dean, J.V, Choe, J. | | Deposit date: | 2017-03-04 | | Release date: | 2017-05-03 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.002 Å) | | Cite: | Differences in salicylic acid glucose conjugations by UGT74F1 and UGT74F2 from Arabidopsis thaliana.
Sci Rep, 7, 2017
|
|
6BSV
 
 | | Crystal structure of Xyloglucan Xylosyltransferase binary form | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, MANGANESE (II) ION, NITRATE ION, ... | | Authors: | Zabotina, O.A, Culbertson, A.T, Ehrlich, J.J, Choe, J, Honzatko, R.B. | | Deposit date: | 2017-12-04 | | Release date: | 2018-05-23 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.433 Å) | | Cite: | Structure of xyloglucan xylosyltransferase 1 reveals simple steric rules that define biological patterns of xyloglucan polymers.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6BSW
 
 | | Crystal structure of Xyloglucan Xylosyltransferase 1 ternary form | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GLYCEROL, MANGANESE (II) ION, ... | | Authors: | Culbertson, A.T, Ehrlich, J.J, Choe, J, Honzatko, R.B, Zabotina, O.A. | | Deposit date: | 2017-12-04 | | Release date: | 2018-05-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.156 Å) | | Cite: | Structure of xyloglucan xylosyltransferase 1 reveals simple steric rules that define biological patterns of xyloglucan polymers.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6BSU
 
 | | Crystal structure of xyloglucan xylosyltransferase I | | Descriptor: | MANGANESE (II) ION, Xyloglucan 6-xylosyltransferase 1 | | Authors: | Culbertson, A.T, Ehrlich, J.J, Choe, J, Honzatko, R.B, Zabotina, O.A. | | Deposit date: | 2017-12-04 | | Release date: | 2018-05-23 | | Last modified: | 2022-03-23 | | Method: | X-RAY DIFFRACTION (1.497 Å) | | Cite: | Structure of xyloglucan xylosyltransferase 1 reveals simple steric rules that define biological patterns of xyloglucan polymers.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
5K2K
 
 | | Crystal structure of lysozyme | | Descriptor: | Lysozyme C | | Authors: | Ko, S, Choe, J. | | Deposit date: | 2016-05-19 | | Release date: | 2017-05-31 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of lysozyme
To Be Published
|
|
5K2Q
 
 | | Lysozyme with nano particles | | Descriptor: | Lysozyme C | | Authors: | Ko, S, Choe, J. | | Deposit date: | 2016-05-19 | | Release date: | 2017-05-31 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Crystal structure of lysozyme with nano particle
To Be Published
|
|
5K2R
 
 | | Crystal structure of lysozyme | | Descriptor: | Lysozyme C | | Authors: | Ko, S, Choe, J. | | Deposit date: | 2016-05-19 | | Release date: | 2017-05-31 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of lysozyme with nano particles
To Be Published
|
|
5K2S
 
 | | Lysozyme with nano particles | | Descriptor: | Lysozyme C | | Authors: | Ko, S, Choe, J. | | Deposit date: | 2016-05-19 | | Release date: | 2017-05-31 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structure of lysozyme with nano particles
To Be Published
|
|
5K2N
 
 | | lysozyme with nano particles | | Descriptor: | Lysozyme C | | Authors: | Ko, S, Choe, J. | | Deposit date: | 2016-05-19 | | Release date: | 2017-05-31 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal Structure of Lysozyme
To Be Published
|
|
5K2P
 
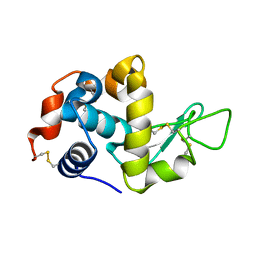 | | Crystal structure of lysozyme | | Descriptor: | Lysozyme C | | Authors: | Ko, S, Choe, J. | | Deposit date: | 2016-05-19 | | Release date: | 2017-05-31 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | Crystal structure of lysozyme
To Be Published
|
|
2R9U
 
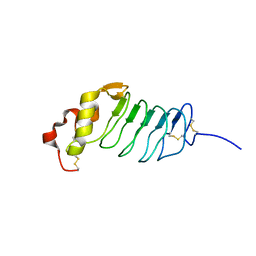 | |
8F5L
 
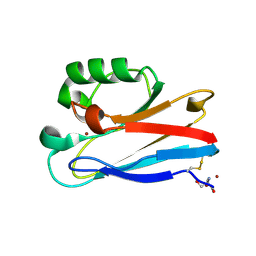 | | Azurin from Pseudomonas aeruginosa, Y72F/Y108F/F110L mutant | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Azurin, COPPER (II) ION | | Authors: | Zeug, M, Offenbacher, A.R, Choe, J. | | Deposit date: | 2022-11-14 | | Release date: | 2023-01-04 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Electrochemical and Structural Study of the Buried Tryptophan in Azurin: Effects of Hydration and Polarity on the Redox Potential of W48.
J.Phys.Chem.B, 127, 2023
|
|
8F5K
 
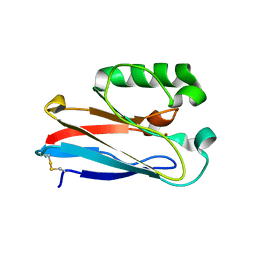 | |
5GJ7
 
 | | putative Acyl-CoA dehydrogenase | | Descriptor: | Acyl-CoA dehydrogenase type 2 domain protein | | Authors: | Moon, H, Choe, J. | | Deposit date: | 2016-06-28 | | Release date: | 2017-07-05 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Putative Acyl CoA dehydrogenase
To Be Published
|
|
2P1I
 
 | | Plasmodium yoelii Ribonucleotide Reductase Subunit R2 (PY03671) | | Descriptor: | FE (III) ION, Ribonucleotide reductase, small chain | | Authors: | Wernimont, A.K, Dong, A, Choe, J, Gao, M, Walker, J, Lew, J, Alam, Z, Zhao, Y, Nordlund, P, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Sundstrom, M, Bochkarev, A, Hui, R, Artz, J.D, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-03-05 | | Release date: | 2007-03-27 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Plasmodium yoelii Ribonucleotide Reductase Subunit R2 (PY03671)
To be Published
|
|
1ZIW
 
 | | Human Toll-like Receptor 3 extracellular domain structure | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, ... | | Authors: | Wilson, I.A, Choe, J. | | Deposit date: | 2005-04-27 | | Release date: | 2005-06-28 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of human toll-like receptor 3 (TLR3) ectodomain.
Science, 309, 2005
|
|
2GHI
 
 | | Crystal Structure of Plasmodium yoelii Multidrug Resistance Protein 2 | | Descriptor: | SULFATE ION, transport protein | | Authors: | Dong, A, Gao, M, Choe, J, Zhao, Y, Lew, J, Wasney, G, Alam, Z, Melone, M, Kozieradzki, I, Vedadi, M, Edwards, A.M, Arrowsmith, C.H, Weigelt, J, Sundstrom, M, Bochkarev, A, Hui, R, Artz, J.D, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-03-27 | | Release date: | 2006-04-25 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Genome-scale protein expression and structural biology of Plasmodium falciparum and related Apicomplexan organisms.
Mol.Biochem.Parasitol., 151, 2007
|
|
1Z6G
 
 | | Crystal structure of guanylate kinase from Plasmodium falciparum | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, SULFATE ION, guanylate kinase | | Authors: | Mulichak, A.M, Lew, J, Artz, J, Choe, J, Walker, J.R, Zhao, Y, Sundstrom, M, Arrowsmith, C, Edwards, A, Bochkarev, A, Hui, R, Gao, M, Structural Genomics Consortium (SGC) | | Deposit date: | 2005-03-22 | | Release date: | 2005-04-05 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Genome-scale protein expression and structural biology of Plasmodium falciparum and related Apicomplexan organisms.
Mol.Biochem.Parasitol., 151, 2007
|
|
1YRV
 
 | | Novel Ubiquitin-Conjugating Enzyme | | Descriptor: | ubiquitin-conjugating ligase MGC351130 | | Authors: | Walker, J.R, Choe, J, Avvakumov, G.V, Newman, E.M, MacKenzie, F, Sundstrom, M, Arrowsmith, C, Edwards, A, Bochkarev, A, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2005-02-04 | | Release date: | 2005-03-22 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | A human ubiquitin conjugating enzyme (E2)-HECT E3 ligase structure-function screen.
Mol Cell Proteomics, 11, 2012
|
|
1ZIV
 
 | | Catalytic Domain of Human Calpain-9 | | Descriptor: | BETA-MERCAPTOETHANOL, CALCIUM ION, Calpain 9 | | Authors: | Walker, J.R, Davis, T, Newman, E.M, Mackenzie, F, Dong, A, Choe, J, Arrowsmith, C, Sundstrom, M, Edwards, A, Bochkarev, A, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2005-04-27 | | Release date: | 2005-07-05 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | The crystal structures of human calpains 1 and 9 imply diverse mechanisms of action and auto-inhibition.
J.Mol.Biol., 366, 2007
|
|
6X9H
 
 | | Molecular mechanism and structural basis of small-molecule modulation of acid-sensing ion channel 1 (ASIC1) | | Descriptor: | 2-[4-(3,4-dimethoxyphenoxy)phenyl]-1H-benzimidazole-6-carboximidamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, Acid-sensing ion channel 1, ... | | Authors: | Liu, Y, Ma, J, DesJarlais, R.L, Hagan, R, Rech, J, Lin, D, Liu, C, Miller, R, Schoellerman, J, Luo, J, Letavic, M, Grasberger, B, Maher, M. | | Deposit date: | 2020-06-02 | | Release date: | 2020-12-30 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | Molecular mechanism and structural basis of small-molecule modulation of the gating of acid-sensing ion channel 1.
Commun Biol, 4, 2021
|
|
8PNL
 
 | | Outward-open conformation of a Major Facilitator Superfamily (MFS) transporter MHAS2168, a homologue of Rv1410 from M. tuberculosis, in complex with an alpaca nanobody | | Descriptor: | Nb_H2, Putative triacylglyceride transporter | | Authors: | Remm, S, Schoeppe, J, Hutter, C.A.J, Gonda, I, Seeger, M.A. | | Deposit date: | 2023-06-30 | | Release date: | 2023-10-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis for triacylglyceride extraction from mycobacterial inner membrane by MFS transporter Rv1410.
Nat Commun, 14, 2023
|
|
