1M66
 
 | | Crystal Structure of Leishmania mexicana GPDH Complexed with Inhibitor 2-bromo-6-chloro-purine | | Descriptor: | 2-BROMO-6-CHLORO-PURINE, Glycerol-3-phosphate dehydrogenase, PALMITIC ACID | | Authors: | Choe, J, Suresh, S, Wisedchaisri, G, Kennedy, K.J, Gelb, M.H, Hol, W.G.J. | | Deposit date: | 2002-07-12 | | Release date: | 2002-12-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Anomalous differences of light elements in determining precise binding modes of ligands to
glycerol-3-phosphate dehydrogenase
Chem.Biol., 9, 2002
|
|
1M67
 
 | | Crystal Structure of Leishmania mexicana GPDH Complexed with Inhibitor 2-bromo-6-hydroxy-purine | | Descriptor: | 2-BROMO-6-HYDROXY-PURINE, Glycerol-3-phosphate dehydrogenase, PALMITIC ACID | | Authors: | Choe, J, Suresh, S, Wisedchaisri, G, Kennedy, K.J, Gelb, M.H, Hol, W.G.J. | | Deposit date: | 2002-07-12 | | Release date: | 2002-12-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Anomalous differences of light elements in determining precise binding modes of ligands to
glycerol-3-phosphate dehydrogenase
Chem.Biol., 9, 2002
|
|
1N1G
 
 | |
1NUW
 
 | | Fructose-1,6-Bisphosphatase Complex with Magnesium, Fructose-6-Phosphate and Phosphate at pH 9.6 | | Descriptor: | 6-O-phosphono-beta-D-fructofuranose, Fructose-1,6-bisphosphatase, MAGNESIUM ION, ... | | Authors: | Choe, J, Iancu, C.V, Fromm, H.J, Honzatko, R.B. | | Deposit date: | 2003-02-01 | | Release date: | 2003-07-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Metaphosphate in the active site of fructose-1,6-bisphosphatase
J.BIOL.CHEM., 278, 2003
|
|
1NUX
 
 | | Fructose-1,6-Bisphosphatase Complex with Magnesium, Fructose-6-Phosphate, Phosphate and inhibitory concentrations of Potassium (200mM) | | Descriptor: | 6-O-phosphono-beta-D-fructofuranose, Fructose-1,6-bisphosphatase, MAGNESIUM ION, ... | | Authors: | Choe, J, Iancu, C.V, Fromm, H.J, Honzatko, R.B. | | Deposit date: | 2003-02-01 | | Release date: | 2003-07-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Metaphosphate in the active site of fructose-1,6-bisphosphatase
J.BIOL.CHEM., 278, 2003
|
|
1NV4
 
 | | Fructose-1,6-Bisphosphatase Complex with Magnesium, Fructose-6-Phosphate, Phosphate, EDTA and Thallium (1 mM) | | Descriptor: | 6-O-phosphono-beta-D-fructofuranose, Fructose-1,6-bisphosphatase, MAGNESIUM ION, ... | | Authors: | Choe, J, Iancu, C.V, Fromm, H.J, Honzatko, R.B. | | Deposit date: | 2003-02-02 | | Release date: | 2003-07-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Interaction of Tl+ with product complexes of fructose-1,6-bisphosphatase
J.BIOL.CHEM., 278, 2003
|
|
1NV0
 
 | | Fructose-1,6-Bisphosphatase Complex with Magnesium, Fructose-6-Phosphate, Phosphate and 1 mM Thallium | | Descriptor: | 6-O-phosphono-beta-D-fructofuranose, Fructose-1,6-bisphosphatase, MAGNESIUM ION, ... | | Authors: | Choe, J, Iancu, C.V, Fromm, H.J, Honzatko, R.B. | | Deposit date: | 2003-02-01 | | Release date: | 2003-07-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Interaction of Tl+ with product complexes of fructose-1,6-bisphosphatase
J.BIOL.CHEM., 278, 2003
|
|
1NV3
 
 | | Fructose-1,6-Bisphosphatase Complex with Magnesium, Fructose-6-Phosphate, Phosphate and Thallium (100 mM) | | Descriptor: | 6-O-phosphono-beta-D-fructofuranose, Fructose-1,6-bisphosphatase, MAGNESIUM ION, ... | | Authors: | Choe, J, Iancu, C.V, Fromm, H.J, Honzatko, R.B. | | Deposit date: | 2003-02-02 | | Release date: | 2003-07-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Interaction of Tl+ with product complexes of fructose-1,6-bisphosphatase
J.BIOL.CHEM., 278, 2003
|
|
1ZO2
 
 | | Structure of nuclear transport factor 2 (Ntf2) from Cryptosporidium parvum | | Descriptor: | nuclear transport factor 2 | | Authors: | Choe, J, Artz, J.D, Gao, M, Lew, J, Zhao, Y, Arrowsmith, C, Edwards, A, Sundstrom, M, Bochkarev, A, Hui, R, Structural Genomics Consortium (SGC) | | Deposit date: | 2005-05-12 | | Release date: | 2005-05-31 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Genome-scale protein expression and structural biology of Plasmodium falciparum and related Apicomplexan organisms.
Mol.Biochem.Parasitol., 151, 2007
|
|
1CNQ
 
 | | FRUCTOSE-1,6-BISPHOSPHATASE COMPLEXED WITH FRUCTOSE-6-PHOSPHATE AND ZINC IONS | | Descriptor: | 6-O-phosphono-beta-D-fructofuranose, FRUCTOSE-1,6-BISPHOSPHATASE, PHOSPHATE ION, ... | | Authors: | Choe, J, Poland, B.W, Fromm, H, Honzatko, R. | | Deposit date: | 1999-05-21 | | Release date: | 1999-05-28 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Role of a dynamic loop in cation activation and allosteric regulation of recombinant porcine fructose-1,6-bisphosphatase.
Biochemistry, 37, 1998
|
|
1EYK
 
 | | FRUCTOSE-1,6-BISPHOSPHATASE COMPLEX WITH AMP, ZINC, FRUCTOSE-6-PHOSPHATE AND PHOSPHATE (T-STATE) | | Descriptor: | 6-O-phosphono-beta-D-fructofuranose, ADENOSINE MONOPHOSPHATE, FRUCTOSE-1,6-BISPHOSPHATASE, ... | | Authors: | Choe, J, Honzatko, R.B. | | Deposit date: | 2000-05-07 | | Release date: | 2000-08-09 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Crystal structures of fructose 1,6-bisphosphatase: mechanism of catalysis and allosteric inhibition revealed in product complexes.
Biochemistry, 39, 2000
|
|
1EYJ
 
 | | FRUCTOSE-1,6-BISPHOSPHATASE COMPLEX WITH AMP, MAGNESIUM, FRUCTOSE-6-PHOSPHATE AND PHOSPHATE (T-STATE) | | Descriptor: | 6-O-phosphono-beta-D-fructofuranose, ADENOSINE MONOPHOSPHATE, FRUCTOSE-1,6-BISPHOSPHATASE, ... | | Authors: | Choe, J, Honzatko, R.B. | | Deposit date: | 2000-05-07 | | Release date: | 2000-08-09 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Crystal structures of fructose 1,6-bisphosphatase: mechanism of catalysis and allosteric inhibition revealed in product complexes.
Biochemistry, 39, 2000
|
|
1EYI
 
 | | FRUCTOSE-1,6-BISPHOSPHATASE COMPLEX WITH MAGNESIUM, FRUCTOSE-6-PHOSPHATE AND PHOSPHATE (R-STATE) | | Descriptor: | 6-O-phosphono-beta-D-fructofuranose, FRUCTOSE-1,6-BISPHOSPHATASE, MAGNESIUM ION, ... | | Authors: | Choe, J, Honzatko, R.B. | | Deposit date: | 2000-05-06 | | Release date: | 2000-08-09 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Crystal structures of fructose 1,6-bisphosphatase: mechanism of catalysis and allosteric inhibition revealed in product complexes.
Biochemistry, 39, 2000
|
|
4NKR
 
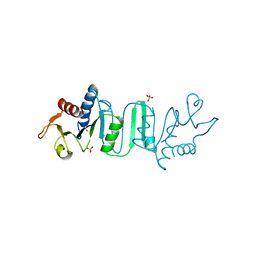 | | The Crystal structure of Bacillus subtilis MobB | | Descriptor: | Molybdopterin-guanine dinucleotide biosynthesis protein B, SULFATE ION | | Authors: | Choe, J, Kim, D, Choi, S, Kim, H. | | Deposit date: | 2013-11-13 | | Release date: | 2014-11-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Insight into the Role of E.coli MobB in Molybdenum Cofactor Biosynthesis based on the high resolution crystal structure
TO BE PUBLISHED
|
|
1YLA
 
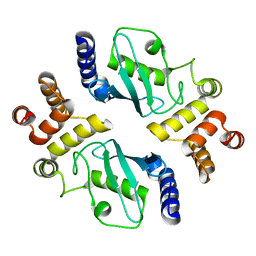 | | Ubiquitin-conjugating enzyme E2-25 kDa (Huntington interacting protein 2) | | Descriptor: | Ubiquitin-conjugating enzyme E2-25 kDa | | Authors: | Choe, J, Avvakumov, G.V, Newman, E.M, Mackenzie, F, Kozieradzki, I, Bochkarev, A, Sundstrom, M, Arrowsmith, C, Edwards, A, Dhe-paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2005-01-19 | | Release date: | 2005-02-01 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis of E2-25K/UBB+1 interaction leading to proteasome inhibition and neurotoxicity
J.Biol.Chem., 285, 2010
|
|
2NLO
 
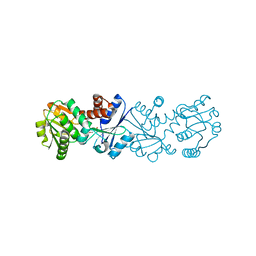 | |
6W54
 
 | | Crystal Structure of Gallic Acid Decarboxylase from Arxula adeninivorans | | Descriptor: | 4-NITROCATECHOL, COBALT (II) ION, Gallate decarboxylase, ... | | Authors: | Zeug, M, Marckovic, N, Iancu, C.V, Tripp, J, Oreb, M, Choe, J. | | Deposit date: | 2020-03-12 | | Release date: | 2021-02-17 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structures of non-oxidative decarboxylases reveal a new mechanism of action with a catalytic dyad and structural twists.
Sci Rep, 11, 2021
|
|
5K2N
 
 | |
5K2P
 
 | | Crystal structure of lysozyme | | Descriptor: | Lysozyme C | | Authors: | Ko, S, Choe, J. | | Deposit date: | 2016-05-19 | | Release date: | 2017-05-31 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | Crystal structure of lysozyme
To Be Published
|
|
5V2J
 
 | | Crystal structure of UDP-glucosyltransferase, UGT74F2 (T15S), with UDP and 2-bromobenzoic acid | | Descriptor: | 2-bromobenzoic acid, UDP-glycosyltransferase 74F2, URIDINE-5'-DIPHOSPHATE, ... | | Authors: | George Thompson, A.M, Iancu, C.V, Dean, J.V, Choe, J. | | Deposit date: | 2017-03-04 | | Release date: | 2017-05-03 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Differences in salicylic acid glucose conjugations by UGT74F1 and UGT74F2 from Arabidopsis thaliana.
Sci Rep, 7, 2017
|
|
5V2K
 
 | | Crystal structure of UDP-glucosyltransferase, UGT74F2 (T15A), with UDP and 2-bromobenzoic acid | | Descriptor: | 2-bromobenzoic acid, UDP-glycosyltransferase 74F2, URIDINE-5'-DIPHOSPHATE, ... | | Authors: | George Thompson, A.M, Iancu, C.V, Dean, J.V, Choe, J. | | Deposit date: | 2017-03-04 | | Release date: | 2017-05-03 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.002 Å) | | Cite: | Differences in salicylic acid glucose conjugations by UGT74F1 and UGT74F2 from Arabidopsis thaliana.
Sci Rep, 7, 2017
|
|
7JMR
 
 | | Crystal structure of the pea pathogenicity protein 2 from Madurella mycetomatis | | Descriptor: | CALCIUM ION, POTASSIUM ION, Pea pathogenicity protein 2 | | Authors: | Zeug, M, Markovic, N, Iancu, C.V, Tripp, J, Oreb, M, Choe, J. | | Deposit date: | 2020-08-02 | | Release date: | 2021-02-17 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Crystal structures of non-oxidative decarboxylases reveal a new mechanism of action with a catalytic dyad and structural twists.
Sci Rep, 11, 2021
|
|
7JMV
 
 | | Crystal structure of the pea pathogenicity protein 2 from Madurella mycetomatis complexed with 4-nitrocatechol | | Descriptor: | 4-NITROCATECHOL, CALCIUM ION, POTASSIUM ION, ... | | Authors: | Zeug, M, Markovic, N, Iancu, C.V, Tripp, J, Oreb, M, Choe, J. | | Deposit date: | 2020-08-03 | | Release date: | 2021-02-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Crystal structures of non-oxidative decarboxylases reveal a new mechanism of action with a catalytic dyad and structural twists.
Sci Rep, 11, 2021
|
|
7KD9
 
 | | Crystal Structure of Gallic Acid Decarboxylase from Arxula adeninivorans | | Descriptor: | Gallate decarboxylase, POTASSIUM ION | | Authors: | Zeug, M, Markovic, N, Iancu, C.V, Tripp, J, Oreb, M, Choe, J. | | Deposit date: | 2020-10-08 | | Release date: | 2021-02-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Crystal structures of non-oxidative decarboxylases reveal a new mechanism of action with a catalytic dyad and structural twists.
Sci Rep, 11, 2021
|
|
5GJ7
 
 | | putative Acyl-CoA dehydrogenase | | Descriptor: | Acyl-CoA dehydrogenase type 2 domain protein | | Authors: | Moon, H, Choe, J. | | Deposit date: | 2016-06-28 | | Release date: | 2017-07-05 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Putative Acyl CoA dehydrogenase
To Be Published
|
|
