4I6K
 
 | | Crystal structure of probable 2-PYRONE-4,6-DICARBOXYLIC ACID HYDROLASE ABAYE1769 (TARGET EFI-505029) from Acinetobacter baumannii with citric acid bound | | Descriptor: | Amidohydrolase family protein, CITRIC ACID | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Zencheck, W.D, Imker, H.J, Al Obaidi, N.F, Stead, M, Love, J, Gerlt, J.A, Raushel, F.M, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-11-29 | | Release date: | 2012-12-12 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.276 Å) | | Cite: | CRYSTAL STRUCTURE OF PROBABLE 2-PYRONE-4,6-DICARBOXYLIC ACID HYDROLASE (TARGET EFI-505029) FROM Acinetobacter baumannii
To be Published
|
|
4ECJ
 
 | | Crystal structure of glutathione s-transferase prk13972 (target efi-501853) from pseudomonas aeruginosa pacs2 complexed with glutathione | | Descriptor: | GLUTATHIONE, glutathione S-transferase | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Zencheck, W.D, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Imker, H.J, Armstrong, R.N, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-03-26 | | Release date: | 2012-04-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Crystal Structure of Glutathione S-Transferase Prk13972 from Pseudomonas Aeruginosa
To be Published
|
|
3G8T
 
 | | Crystal structure of the G33A mutant Bacillus anthracis glmS ribozyme bound to GlcN6P | | Descriptor: | 2-amino-2-deoxy-6-O-phosphono-alpha-D-glucopyranose, MAGNESIUM ION, RNA (5'-R(*AP*(A2M)P*GP*CP*GP*CP*CP*AP*GP*AP*AP*CP*U)-3'), ... | | Authors: | Strobel, S.A, Cochrane, J.C, Lipchock, S.V, Smith, K.D. | | Deposit date: | 2009-02-12 | | Release date: | 2009-11-03 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural and chemical basis for glucosamine 6-phosphate binding and activation of the glmS ribozyme
Biochemistry, 48, 2009
|
|
4EXJ
 
 | | Crystal structure of glutathione s-transferase like protein lelg_03239 (target efi-501752) from lodderomyces elongisporus | | Descriptor: | CHLORIDE ION, SULFATE ION, uncharacterized protein | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Zencheck, W.D, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Imker, H.J, Armstrong, R.N, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-04-30 | | Release date: | 2012-05-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Crystal structure of glutathione s-transferase like protein lelg_03239 (target efi-501752) from lodderomyces elongisporus
To be Published
|
|
2LOM
 
 | | Backbone structure of human membrane protein HIGD1A | | Descriptor: | HIG1 domain family member 1A | | Authors: | Blain, K, Klammt, C, Maslennikov, I, Kwiatkowski, W, Choe, S. | | Deposit date: | 2012-01-26 | | Release date: | 2012-05-23 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Facile backbone structure determination of human membrane proteins by NMR spectroscopy.
Nat.Methods, 9, 2012
|
|
2LOS
 
 | | Backbone structure of human membrane protein TMEM14C | | Descriptor: | Transmembrane protein 14C | | Authors: | Klammt, C, Vajpai, N, Maslennikov, I, Riek, R, Choe, S. | | Deposit date: | 2012-01-26 | | Release date: | 2012-05-23 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Facile backbone structure determination of human membrane proteins by NMR spectroscopy.
Nat.Methods, 9, 2012
|
|
3H8B
 
 | | A combined crystallographic and molecular dynamics study of cathepsin-L retro-binding inhibitors(compound 9) | | Descriptor: | Cathepsin L1, N~2~,N~6~-bis(biphenyl-4-ylacetyl)-L-lysyl-D-arginyl-N-(2-phenylethyl)-L-phenylalaninamide | | Authors: | Tulsidas, S.R, Chowdhury, S.F, Kumar, S, Joseph, L, Purisima, E.O, Sivaraman, J. | | Deposit date: | 2009-04-29 | | Release date: | 2009-10-20 | | Last modified: | 2014-02-05 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A combined crystallographic and molecular dynamics study of cathepsin L retrobinding inhibitors
J.Med.Chem., 52, 2009
|
|
2LOQ
 
 | | Backbone structure of human membrane protein FAM14B (Interferon alpha-inducible protein 27-like protein 1) | | Descriptor: | Interferon alpha-inducible protein 27-like protein 1 | | Authors: | Klammt, C, Chui, E.J, Maslennikov, I, Kwiatkowski, W, Choe, S. | | Deposit date: | 2012-01-26 | | Release date: | 2012-05-23 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Facile backbone structure determination of human membrane proteins by NMR spectroscopy.
Nat.Methods, 9, 2012
|
|
3H89
 
 | | A combined crystallographic and molecular dynamics study of cathepsin-L retro-binding inhibitors(compound 4) | | Descriptor: | Cathepsin L1, N~2~,N~6~-bis(biphenyl-4-ylacetyl)-L-lysyl-D-arginyl-N-(2-phenylethyl)-L-tyrosinamide | | Authors: | Tulsidas, S.R, Chowdhury, S.F, Kumar, S, Joseph, L, Purisima, E.O, Sivaraman, J. | | Deposit date: | 2009-04-29 | | Release date: | 2009-10-20 | | Last modified: | 2014-02-05 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A combined crystallographic and molecular dynamics study of cathepsin L retrobinding inhibitors
J.Med.Chem., 52, 2009
|
|
3E4A
 
 | | Human IDE-inhibitor complex at 2.6 angstrom resolution | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, ACETIC ACID, HYDROXAMATE PEPTIDE II1, ... | | Authors: | Malito, E, Leissring, M.A, Choi, S, Cuny, G.D, Tang, W.J. | | Deposit date: | 2008-08-11 | | Release date: | 2009-05-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Designed inhibitors of insulin-degrading enzyme regulate the catabolism and activity of insulin.
Plos One, 5, 2010
|
|
6JJC
 
 | | Cryo-EM structure of giant freshwater prawn Macrobrachium rosenbergii nodavirus (MrNV) semi-empty VLP | | Descriptor: | CALCIUM ION, Capsid protein | | Authors: | Chang, W.H, Wang, C.H, Lin, H.H, Lin, S.Y, Chong, S.C, Wu, Y.Y. | | Deposit date: | 2019-02-25 | | Release date: | 2019-05-01 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.92 Å) | | Cite: | Cryo-EM structure of giant freshwater prawn Macrobrachium rosenbergii nodavirus (MrNV) semi-empty VLP
To Be Published
|
|
6BZH
 
 | | Structure of mouse RIG-I tandem CARDs | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Probable ATP-dependent RNA helicase DDX58 | | Authors: | Kershaw, N.J, D'Cruz, A.A, Babon, J.J, Nicholson, S.E. | | Deposit date: | 2017-12-24 | | Release date: | 2018-01-17 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Identification of a second binding site on the TRIM25 B30.2 domain.
Biochem. J., 475, 2018
|
|
4P9K
 
 | | CRYSTAL STRUCTURE OF A TRAP PERIPLASMIC SOLUTE BINDING PROTEIN FROM VERMINEPHROBACTER EISENIAE EF01-2 (Veis_3954, TARGET EFI-510324) A NEPHRIDIAL SYMBIONT OF THE EARTHWORM EISENIA FOETIDA, BOUND TO D-ERYTHRONATE WITH RESIDUAL DENSITY SUGGESTIVE OF SUPERPOSITION WITH COPURIFIED ALTERNATIVE LIGAND. | | Descriptor: | (2R,3R)-2,3,4-trihydroxybutanoic acid, TRAP dicarboxylate transporter, DctP subunit | | Authors: | Vetting, M.W, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Stead, M, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Evans, B, Hillerich, B, Love, J, Seidel, R.D, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2014-04-04 | | Release date: | 2014-05-07 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Experimental strategies for functional annotation and metabolism discovery: targeted screening of solute binding proteins and unbiased panning of metabolomes.
Biochemistry, 54, 2015
|
|
4PAI
 
 | | CRYSTAL STRUCTURE OF A TRAP PERIPLASMIC SOLUTE BINDING PROTEIN FROM RUEGERIA POMEROYI DSS-3 (SPO1773, TARGET EFI-510260) WITH BOUND 3-HYDROXYBENZOATE | | Descriptor: | 3-HYDROXYBENZOIC ACID, TRAP dicarboxylate transporter, DctP subunit, ... | | Authors: | Vetting, M.W, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Stead, M, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Evans, B, Hillerich, B, Love, J, Seidel, R.D, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2014-04-09 | | Release date: | 2014-05-07 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Experimental strategies for functional annotation and metabolism discovery: targeted screening of solute binding proteins and unbiased panning of metabolomes.
Biochemistry, 54, 2015
|
|
1A68
 
 | |
4PAF
 
 | | CRYSTAL STRUCTURE OF A TRAP PERIPLASMIC SOLUTE BINDING PROTEIN FROM RUEGERIA POMEROYI DSS-3 (SPO1773, TARGET EFI-510260) WITH BOUND 3,4-DIHYDROXYBENZOATE | | Descriptor: | 3,4-DIHYDROXYBENZOIC ACID, MALONATE ION, TRAP dicarboxylate transporter, ... | | Authors: | Vetting, M.W, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Stead, M, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Evans, B, Hillerich, B, Love, J, Seidel, R.D, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2014-04-08 | | Release date: | 2014-05-07 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Experimental strategies for functional annotation and metabolism discovery: targeted screening of solute binding proteins and unbiased panning of metabolomes.
Biochemistry, 54, 2015
|
|
4PBH
 
 | | CRYSTAL STRUCTURE OF A TRAP PERIPLASMIC SOLUTE BINDING PROTEIN FROM RUEGERIA POMEROYI DSS-3 (SPO1773, TARGET EFI-510260) WITH BOUND BENZOIC ACID | | Descriptor: | BENZOIC ACID, TRAP dicarboxylate transporter, DctP subunit, ... | | Authors: | Vetting, M.W, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Stead, M, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Evans, B, Hillerich, B, Love, J, Seidel, R.D, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2014-04-12 | | Release date: | 2014-05-07 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Experimental strategies for functional annotation and metabolism discovery: targeted screening of solute binding proteins and unbiased panning of metabolomes.
Biochemistry, 54, 2015
|
|
4ZJ1
 
 | | Crystal Structure of p-acrylamido-phenylalanine modified TEM1 beta-lactamase from Escherichia coli : V216AcrF mutant | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Beta-lactamase TEM, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Xiao, H, Nasertorabi, F, Choi, S, Han, G.W, Reed, S.A, Stevens, R.C, Schultz, P.G. | | Deposit date: | 2015-04-28 | | Release date: | 2015-05-20 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Exploring the potential impact of an expanded genetic code on protein function.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
1B8K
 
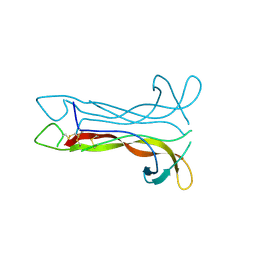 | | Neurotrophin-3 from Human | | Descriptor: | PROTEIN (NEUROTROPHIN-3) | | Authors: | Robinson, R.C, Radziejewski, C, Stuart, D.I, Jones, E.Y, Choe, S. | | Deposit date: | 1999-02-01 | | Release date: | 1999-02-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The structures of the neurotrophin 4 homodimer and the brain-derived neurotrophic factor/neurotrophin 4 heterodimer reveal a common Trk-binding site.
Protein Sci., 8, 1999
|
|
4GLT
 
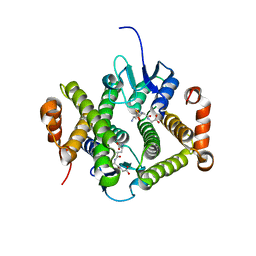 | | Crystal structure of glutathione s-transferase MFLA_2116 (target EFI-507160) from methylobacillus flagellatus kt with gsh bound | | Descriptor: | GLUTATHIONE, Glutathione S-transferase-like protein | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Zencheck, W.D, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Imker, H.J, Armstrong, R.N, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-08-14 | | Release date: | 2012-08-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of glutathione s-transferase MFLA_2116 (target EFI-507160) from methylobacillus flagellatus kt with gsh bound
To be Published
|
|
7V5C
 
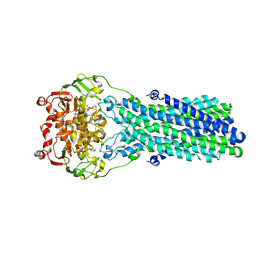 | | Cryo-EM structure of the mouse ABCB9 (ADP.BeF3-bound) | | Descriptor: | ABC-type oligopeptide transporter ABCB9, ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, ... | | Authors: | Park, J.G, Kim, S, Jang, E, Choi, S.H, Han, H, Kim, J.W, Ju, S, Min, D.S, Jin, M.S. | | Deposit date: | 2021-08-17 | | Release date: | 2022-10-19 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | The lysosomal transporter TAPL has a dual role as peptide translocator and phosphatidylserine floppase.
Nat Commun, 13, 2022
|
|
7V5D
 
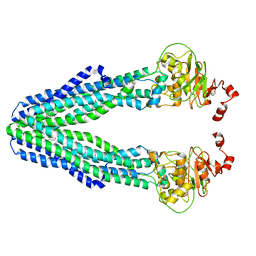 | | Cryo-EM structure of the mouse ABCB9 (PG-bound) | | Descriptor: | (1S)-2-{[{[(2R)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL STEARATE, ABC-type oligopeptide transporter ABCB9 | | Authors: | Park, J.G, Kim, S, Jang, E, Choi, S.H, Han, H, Ju, S, Kim, J.W, Min, D.S, Jin, M.S. | | Deposit date: | 2021-08-17 | | Release date: | 2022-10-19 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | The lysosomal transporter TAPL has a dual role as peptide translocator and phosphatidylserine floppase.
Nat Commun, 13, 2022
|
|
4ZJ3
 
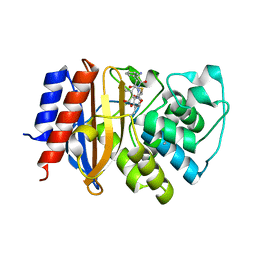 | | Crystal structure of cephalexin bound acyl-enzyme intermediate of Val216AcrF mutant TEM1 beta-lactamase from Escherichia coli: E166N and V216AcrF mutant. | | Descriptor: | Beta-lactamase TEM | | Authors: | Xiao, H, Nasertorabi, F, Choi, S, Han, G.W, Reed, S.A, Stevens, R.C, Schultz, P.G. | | Deposit date: | 2015-04-29 | | Release date: | 2015-05-20 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Exploring the potential impact of an expanded genetic code on protein function.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
1B98
 
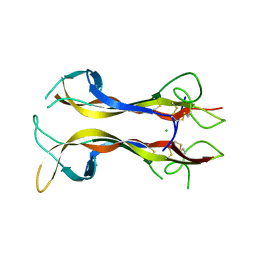 | | NEUROTROPHIN 4 (HOMODIMER) | | Descriptor: | CHLORIDE ION, PROTEIN (NEUROTROPHIN-4) | | Authors: | Robinson, R.C, Radziejewski, C, Stuart, D.I, Jones, E.Y, Choe, S. | | Deposit date: | 1999-02-22 | | Release date: | 1999-02-26 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | The structures of the neurotrophin 4 homodimer and the brain-derived neurotrophic factor/neurotrophin 4 heterodimer reveal a common Trk-binding site.
Protein Sci., 8, 1999
|
|
4DEJ
 
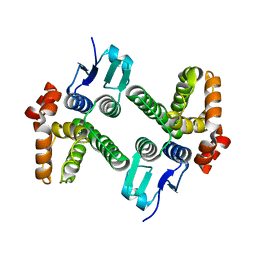 | | Crystal structure of glutathione transferase-like protein IL0419 (Target EFI-501089) from Idiomarina loihiensis L2TR | | Descriptor: | Glutathione S-transferase related protein | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Zencheck, W.D, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Imker, H.J, Armstrong, R.N, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-01-20 | | Release date: | 2012-02-01 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal Structure of Glutathione S-Transferase-Like Protein Il0419 from Idiomarina Loihiensis
To be Published
|
|
