7X9R
 
 | | Crystal structure of the antirepressor GmaR | | Descriptor: | Glycosyl transferase family 2 | | Authors: | Cho, S.Y, Na, H.W, Oh, H.B, Kwak, Y.M, Song, W.S, Park, S.C, Yoon, S.I. | | Deposit date: | 2022-03-16 | | Release date: | 2022-11-09 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural basis of flagellar motility regulation by the MogR repressor and the GmaR antirepressor in Listeria monocytogenes.
Nucleic Acids Res., 50, 2022
|
|
7X9S
 
 | | Crystal structure of a complex between the antirepressor GmaR and the transcriptional repressor MogR | | Descriptor: | GmaR, Motility gene repressor MogR | | Authors: | Cho, S.Y, Na, H.W, Oh, H.B, Kwak, Y.M, Song, W.S, Park, S.C, Yoon, S.I. | | Deposit date: | 2022-03-16 | | Release date: | 2022-11-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.11 Å) | | Cite: | Structural basis of flagellar motility regulation by the MogR repressor and the GmaR antirepressor in Listeria monocytogenes.
Nucleic Acids Res., 50, 2022
|
|
8J56
 
 | | Crystal structure of the FlhDC complex from Cupriavidus necator | | Descriptor: | Flagellar transcriptional regulator FlhC, Flagellar transcriptional regulator FlhD, ZINC ION | | Authors: | Cho, S.Y, Oh, H.B, Yoon, S.I. | | Deposit date: | 2023-04-21 | | Release date: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Hexameric structure of the flagellar master regulator FlhDC from Cupriavidus necator and its interaction with flagellar promoter DNA.
Biochem.Biophys.Res.Commun., 672, 2023
|
|
5YSO
 
 | | Crystal structure of Estrogen Related Receptor-3 (ERR-gamma) ligand binding domain with DN200434 | | Descriptor: | 4-[5-oxidanyl-2-phenyl-1-[4-(4-propan-2-ylpiperazin-1-yl)phenyl]pent-1-enyl]phenol, Estrogen-related receptor gamma | | Authors: | Cho, S.J, Chin, J.W, Yoon, H.S, Jeon, Y.H, Bae, J.H, Song, J.Y. | | Deposit date: | 2017-11-14 | | Release date: | 2018-12-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.501 Å) | | Cite: | A Novel Orally Active Inverse Agonist of Estrogen-related Receptor Gamma (ERR gamma ), DN200434, A Booster of NIS in Anaplastic Thyroid Cancer.
Clin.Cancer Res., 25, 2019
|
|
5XLJ
 
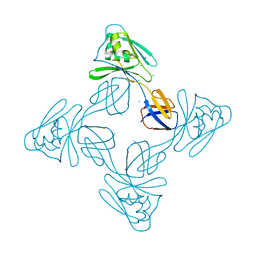 | | Crystal structure of the flagellar cap protein flid D2-D3 domains from serratia marcescens in Space group P432 | | Descriptor: | CHLORIDE ION, Flagellar hook-associated protein 2, SODIUM ION | | Authors: | Cho, S.Y, Song, W.S, Hong, H.J, Yoon, S.I. | | Deposit date: | 2017-05-10 | | Release date: | 2017-06-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Tetrameric structure of the flagellar cap protein FliD from Serratia marcescens.
Biochem. Biophys. Res. Commun., 489, 2017
|
|
5XLK
 
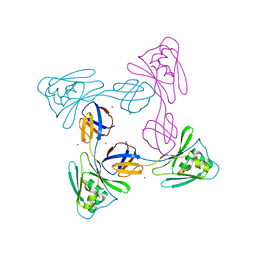 | | Crystal structure of the flagellar cap protein FliD D2-D3 domains from Serratia marcescens in Space group I422 | | Descriptor: | Flagellar hook-associated protein 2, ZINC ION | | Authors: | Cho, S.Y, Song, W.S, Hong, H.J, Yoon, S.I. | | Deposit date: | 2017-05-10 | | Release date: | 2017-06-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Tetrameric structure of the flagellar cap protein FliD from Serratia marcescens.
Biochem. Biophys. Res. Commun., 489, 2017
|
|
6IWY
 
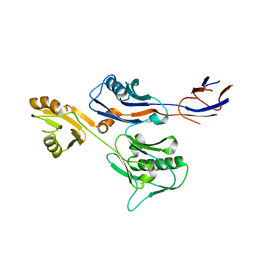 | |
4NQ3
 
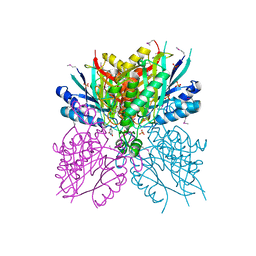 | | Crystal structure of cyanuic acid hydrolase from A. caulinodans | | Descriptor: | BARBITURIC ACID, Cyanuric acid amidohydrolase, MAGNESIUM ION, ... | | Authors: | Cho, S, Shi, K, Aihara, H. | | Deposit date: | 2013-11-23 | | Release date: | 2014-09-10 | | Method: | X-RAY DIFFRACTION (2.702 Å) | | Cite: | Cyanuric acid hydrolase from Azorhizobium caulinodans ORS 571: crystal structure and insights into a new class of Ser-Lys dyad proteins.
Plos One, 9, 2014
|
|
6KTY
 
 | |
6M58
 
 | |
3G8K
 
 | | Crystal structure of murine natural killer cell receptor, Ly49L4 | | Descriptor: | Lectin-related NK cell receptor LY49L1 | | Authors: | Cho, S. | | Deposit date: | 2009-02-12 | | Release date: | 2009-11-17 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Distinct conformations of Ly49 natural killer cell receptors mediate MHC class I recognition in trans and cis.
Immunity, 31, 2009
|
|
3G8L
 
 | | Crystal structure of murine natural killer cell receptor, Ly49L4 | | Descriptor: | Lectin-related NK cell receptor LY49L1 | | Authors: | Cho, S. | | Deposit date: | 2009-02-12 | | Release date: | 2009-11-17 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Distinct conformations of Ly49 natural killer cell receptors mediate MHC class I recognition in trans and cis.
Immunity, 31, 2009
|
|
1IB4
 
 | |
1K81
 
 | |
2EAV
 
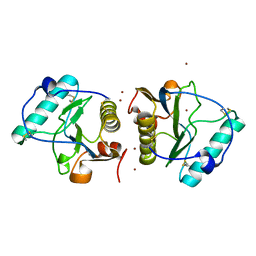 | |
2EAX
 
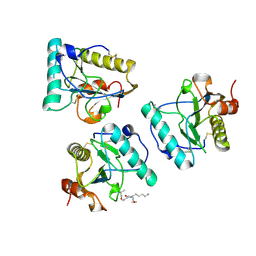 | | Crystal structure of human PGRP-IBETAC in complex with glycosamyl muramyl pentapeptide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-methyl 2-acetamido-3-O-[(1R)-1-carboxyethyl]-2-deoxy-beta-D-glucopyranoside, GLYCOSAMYL MURAMYL PENTAPEPTIDE, Peptidoglycan recognition protein-I-beta | | Authors: | Cho, S. | | Deposit date: | 2007-02-03 | | Release date: | 2007-10-02 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural insights into the bactericidal mechanism of human peptidoglycan recognition proteins
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
2AQ2
 
 | | Crystal structure of T-cell receptor V beta domain variant complexed with superantigen SEC3 mutant | | Descriptor: | Enterotoxin type C-3, SODIUM ION, SULFATE ION, ... | | Authors: | Cho, S, Swaminathan, C.P, Yang, J, Kerzic, M.C, Guan, R, Kieke, M.C, Kranz, D.M, Mariuzza, R.A, Sundberg, E.J. | | Deposit date: | 2005-08-17 | | Release date: | 2006-03-21 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis of affinity maturation and intramolecular cooperativity in a protein-protein interaction.
Structure, 13, 2005
|
|
1K8B
 
 | |
2APW
 
 | | Crystal Structure of the G17E/A52V/S54N/K66E/E80V/L81S/T87S/G96V variant of the murine T cell receptor V beta 8.2 domain | | Descriptor: | MALONIC ACID, T cell receptor beta chain V | | Authors: | Cho, S, Swaminathan, C.P, Yang, J, Kerzic, M.C, Guan, R, Kieke, M.C, Kranz, D.M, Mariuzza, R.A, Sundberg, E.J. | | Deposit date: | 2005-08-16 | | Release date: | 2006-03-21 | | Last modified: | 2018-04-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis of affinity maturation and intramolecular cooperativity in a protein-protein interaction.
Structure, 13, 2005
|
|
2APV
 
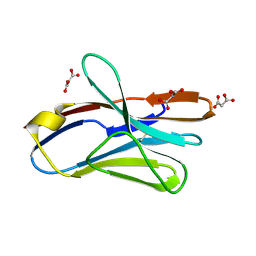 | | Crystal Structure of the G17E/A52V/S54N/Q72H/E80V/L81S/T87S/G96V variant of the murine T cell receptor V beta 8.2 domain | | Descriptor: | MALONIC ACID, T cell receptor beta chain V | | Authors: | Cho, S, Swaminathan, C.P, Yang, J, Kerzic, M.C, Guan, R, Kieke, M.C, Kranz, D.M, Mariuzza, R.A, Sundberg, E.J. | | Deposit date: | 2005-08-16 | | Release date: | 2006-03-21 | | Last modified: | 2018-04-04 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis of affinity maturation and intramolecular cooperativity in a protein-protein interaction.
Structure, 13, 2005
|
|
2AQ1
 
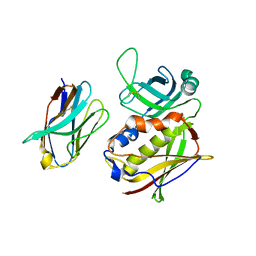 | | Crystal structure of T-cell receptor V beta domain variant complexed with superantigen SEC3 mutant | | Descriptor: | Enterotoxin type C-3, T-cell receptor beta chain V | | Authors: | Cho, S, Swaminathan, C.P, Yang, J, Kerzic, M.C, Guan, R, Kieke, M.C, Kranz, D.M, Mariuzza, R.A, Sundberg, E.J. | | Deposit date: | 2005-08-17 | | Release date: | 2006-03-21 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis of affinity maturation and intramolecular cooperativity in a protein-protein interaction.
Structure, 13, 2005
|
|
2APF
 
 | | Crystal Structure of the A52V/S54N/K66E variant of the murine T cell receptor V beta 8.2 domain | | Descriptor: | MALONIC ACID, T cell receptor beta chain V | | Authors: | Cho, S, Swaminathan, C.P, Yang, J, Kerzic, M.C, Guan, R, Kieke, M.C, Kranz, D.M, Mariuzza, R.A, Sundberg, E.J. | | Deposit date: | 2005-08-16 | | Release date: | 2006-03-21 | | Last modified: | 2018-04-04 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis of affinity maturation and intramolecular cooperativity in a protein-protein interaction.
Structure, 13, 2005
|
|
2APT
 
 | | Crystal Structure of the G17E/S54N/K66E/Q72H/E80V/L81S/T87S/G96V variant of the murine T cell receptor V beta 8.2 domain | | Descriptor: | MALONIC ACID, T-cell receptor beta chain V | | Authors: | Cho, S, Swaminathan, C.P, Yang, J, Kerzic, M.C, Guan, R, Kieke, M.C, Kranz, D.M, Mariuzza, R.A, Sundberg, E.J. | | Deposit date: | 2005-08-16 | | Release date: | 2006-03-21 | | Last modified: | 2018-04-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis of affinity maturation and intramolecular cooperativity in a protein-protein interaction.
Structure, 13, 2005
|
|
2AQ3
 
 | | Crystal structure of T-cell receptor V beta domain variant complexed with superantigen SEC3 | | Descriptor: | Enterotoxin type C-3, T-cell receptor beta chain V | | Authors: | Cho, S, Swaminathan, C.P, Yang, J, Kerzic, M.C, Guan, R, Kieke, M.C, Kranz, D.M, Mariuzza, R.A, Sundberg, E.J. | | Deposit date: | 2005-08-17 | | Release date: | 2006-03-21 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis of affinity maturation and intramolecular cooperativity in a protein-protein interaction.
Structure, 13, 2005
|
|
2APB
 
 | | Crystal Structure of the S54N variant of murine T cell receptor Vbeta 8.2 domain | | Descriptor: | MALONIC ACID, T-cell receptor beta chain V | | Authors: | Cho, S, Swaminathan, C.P, Yang, J, Kerzic, M.C, Guan, R, Kieke, M.C, Kranz, D.M, Mariuzza, R.A, Sundberg, E.J. | | Deposit date: | 2005-08-16 | | Release date: | 2006-03-21 | | Last modified: | 2018-04-04 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis of affinity maturation and intramolecular cooperativity in a protein-protein interaction.
Structure, 13, 2005
|
|
