5BMU
 
 | |
6KQQ
 
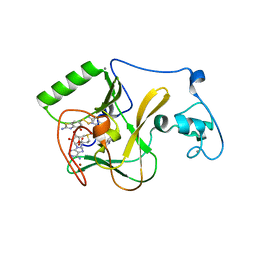 | | NSD1 SET domain in complex with BT3 and SAM | | Descriptor: | 2-azanyl-6-[(2-azanyl-4-oxidanyl-1,3-benzothiazol-6-yl)disulfanyl]-1,3-benzothiazol-4-ol, 2-azanyl-6-sulfanyl-1,3-benzothiazol-4-ol, CALCIUM ION, ... | | Authors: | Cho, H.J, Cierpicki, T. | | Deposit date: | 2019-08-18 | | Release date: | 2020-09-02 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Covalent inhibition of NSD1 histone methyltransferase.
Nat.Chem.Biol., 16, 2020
|
|
6KQP
 
 | | NSD1 SET domain in complex with SAM | | Descriptor: | Histone-lysine N-methyltransferase, H3 lysine-36 and H4 lysine-20 specific, S-ADENOSYLMETHIONINE, ... | | Authors: | Cho, H.J, Cierpicki, T. | | Deposit date: | 2019-08-18 | | Release date: | 2020-09-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Covalent inhibition of NSD1 histone methyltransferase.
Nat.Chem.Biol., 16, 2020
|
|
6WI7
 
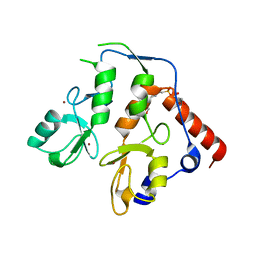 | | RING1B-BMI1 fusion in closed conformation | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, E3 ubiquitin-protein ligase RING2, Polycomb complex protein BMI-1 chimera, ... | | Authors: | Cho, H.J, Cierpicki, T. | | Deposit date: | 2020-04-08 | | Release date: | 2021-04-14 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.702 Å) | | Cite: | Small-molecule inhibitors targeting Polycomb repressive complex 1 RING domain.
Nat.Chem.Biol., 17, 2021
|
|
6WI8
 
 | |
5VNA
 
 | | Crystal structure of human YEATS domain | | Descriptor: | 1,2-ETHANEDIOL, 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, SULFATE ION, ... | | Authors: | Cho, H.J, Cierpicki, T. | | Deposit date: | 2017-04-29 | | Release date: | 2018-09-05 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | GAS41 Recognizes Diacetylated Histone H3 through a Bivalent Binding Mode.
ACS Chem. Biol., 13, 2018
|
|
5VNB
 
 | | YEATS in complex with histone H3 | | Descriptor: | 1,2-ETHANEDIOL, H3K23acK27ac peptide, SULFATE ION, ... | | Authors: | Cho, H.J, Cierpicki, T. | | Deposit date: | 2017-04-29 | | Release date: | 2018-09-05 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | GAS41 Recognizes Diacetylated Histone H3 through a Bivalent Binding Mode.
ACS Chem. Biol., 13, 2018
|
|
5YZH
 
 | | Crystal Structure of Mouse Cytosolic Isocitrate Dehydrogenase | | Descriptor: | GLYCEROL, Isocitrate dehydrogenase [NADP] cytoplasmic, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Cho, H.J, Kang, B.S. | | Deposit date: | 2017-12-14 | | Release date: | 2018-07-04 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.994 Å) | | Cite: | NADP+-dependent cytosolic isocitrate dehydrogenase provides NADPH in the presence of cadmium due to the moderate chelating effect of glutathione.
J. Biol. Inorg. Chem., 23, 2018
|
|
5YZI
 
 | | Crystal Structure of Mouse Cytosolic Isocitrate Dehydrogenase complexed with Cadmium | | Descriptor: | CADMIUM ION, Isocitrate dehydrogenase [NADP] cytoplasmic, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Cho, H.J, Kang, B.S. | | Deposit date: | 2017-12-14 | | Release date: | 2018-07-04 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.519 Å) | | Cite: | NADP+-dependent cytosolic isocitrate dehydrogenase provides NADPH in the presence of cadmium due to the moderate chelating effect of glutathione.
J. Biol. Inorg. Chem., 23, 2018
|
|
7XKI
 
 | | Human Cx36/GJD2 (N-terminal deletion BRIL-fused mutant) gap junction channel in soybean lipids (D6 symmetry) | | Descriptor: | 1,2-DIMYRISTOYL-RAC-GLYCERO-3-PHOSPHOCHOLINE, Gap junction delta-2 protein,Soluble cytochrome b562 | | Authors: | Cho, H.J, Lee, S.N, Jeong, H, Ryu, B, Lee, H.J, Woo, J.S, Lee, H.H. | | Deposit date: | 2022-04-19 | | Release date: | 2023-03-22 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-EM structures of human Cx36/GJD2 neuronal gap junction channel.
Nat Commun, 14, 2023
|
|
7XKT
 
 | | Human Cx36/GJD2 (BRIL-fused mutant) gap junction channel in detergents at 2.2 Angstroms resolution | | Descriptor: | 1,2-DIMYRISTOYL-RAC-GLYCERO-3-PHOSPHOCHOLINE, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, CHOLESTEROL HEMISUCCINATE, ... | | Authors: | Cho, H.J, Lee, S.N, Jeong, H, Ryu, B, Lee, H.J, Woo, J.S, Lee, H.H. | | Deposit date: | 2022-04-20 | | Release date: | 2023-03-22 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.2 Å) | | Cite: | Cryo-EM structures of human Cx36/GJD2 neuronal gap junction channel.
Nat Commun, 14, 2023
|
|
2WAW
 
 | |
2WEE
 
 | |
2WE9
 
 | |
8XH8
 
 | |
8XGG
 
 | | Human Cx36/GJD2 gap junction channel in complex with 1-hexanol. | | Descriptor: | 1,2-DIMYRISTOYL-RAC-GLYCERO-3-PHOSPHOCHOLINE, Gap junction delta-2 protein, HEXAN-1-OL | | Authors: | Cho, H.J, Lee, H.H. | | Deposit date: | 2023-12-15 | | Release date: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Mefloquine-induced conformational shift in Cx36 N-terminal helix leading to channel closure mediated by lipid bilayer.
Nat Commun, 15, 2024
|
|
8XGJ
 
 | | Human Cx36/GJD2 gap junction channel in complex with mefloquine. | | Descriptor: | 1,2-DIMYRISTOYL-RAC-GLYCERO-3-PHOSPHOCHOLINE, Gap junction delta-2 protein, Mefloquine | | Authors: | Cho, H.J, Lee, H.H. | | Deposit date: | 2023-12-15 | | Release date: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Mefloquine-induced conformational shift in Cx36 N-terminal helix leading to channel closure mediated by lipid bilayer.
Nat Commun, 15, 2024
|
|
8XGF
 
 | |
8XGE
 
 | | Human Cx36/GJD2 gap junction channel in porcine brain lipids. | | Descriptor: | 1,2-DIMYRISTOYL-RAC-GLYCERO-3-PHOSPHOCHOLINE, Gap junction delta-2 protein | | Authors: | Cho, H.J, Lee, H.H. | | Deposit date: | 2023-12-15 | | Release date: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.89 Å) | | Cite: | Mefloquine-induced conformational shift in Cx36 N-terminal helix leading to channel closure mediated by lipid bilayer.
Nat Commun, 15, 2024
|
|
8XGD
 
 | |
8XH9
 
 | |
5AIZ
 
 | | The PIAS-like coactivator Zmiz1 is a direct and selective cofactor of Notch1 in T-cell development and leukemia | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, UNKNOWN ATOM OR ION, ... | | Authors: | Cho, H.J, Murai, M, Chiang, M, Cierpicki, T. | | Deposit date: | 2015-02-18 | | Release date: | 2015-10-28 | | Last modified: | 2015-12-02 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Pias-Like Coactivator Zmiz1 is a Direct and Selective Cofactor of Notch1 in T-Cell Development and Leukemia
Immunity, 43, 2015
|
|
2WL9
 
 | | Crystal structure of catechol 2,3-dioxygenase | | Descriptor: | 3-METHYLCATECHOL, CATECHOL 2,3-DIOXYGENASE, FE (III) ION, ... | | Authors: | Cho, H.J, Kim, K.J, Kang, B.S. | | Deposit date: | 2009-06-23 | | Release date: | 2010-09-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Substrate-Binding Mechanism of a Type I Extradiol Dioxygenase.
J.Biol.Chem., 285, 2010
|
|
2XVJ
 
 | |
7C0J
 
 | | Crystal structure of chimeric mutant of GH5 in complex with Z-DNA | | Descriptor: | DNA (5'-D(*TP*CP*GP*CP*GP*CP*G)-3'), Histone H5,Double-stranded RNA-specific adenosine deaminase | | Authors: | Choi, H.J, Park, C.H. | | Deposit date: | 2020-05-01 | | Release date: | 2020-12-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Dual conformational recognition by Z-DNA binding protein is important for the B-Z transition process.
Nucleic Acids Res., 48, 2020
|
|
