5YB9
 
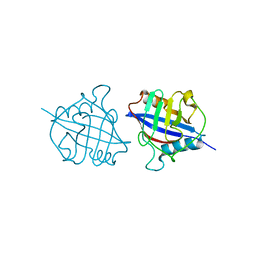 | | Crystal structure of a dimeric cyclophilin A from T.vaginalis | | Descriptor: | Peptidyl-prolyl cis-trans isomerase | | Authors: | Cho, C.C, Lin, M.H, Chou, C.C, Martin, T, Chen, C, Hsu, C.H. | | Deposit date: | 2017-09-04 | | Release date: | 2018-07-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.276 Å) | | Cite: | Structural basis of interaction between dimeric cyclophilin 1 and Myb1 transcription factor in Trichomonas vaginalis
Sci Rep, 8, 2018
|
|
5YBA
 
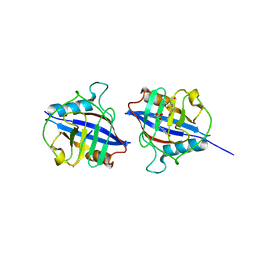 | | Dimeric Cyclophilin from T.vaginalis in complex with Myb1 peptide | | Descriptor: | Myb1 peptide, Peptidyl-prolyl cis-trans isomerase | | Authors: | Cho, C.C, Lin, M.H, Martin, T, Chou, C.C, Chen, C, Hsu, C.H. | | Deposit date: | 2017-09-04 | | Release date: | 2018-07-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.062 Å) | | Cite: | Structural basis of interaction between dimeric cyclophilin 1 and Myb1 transcription factor in Trichomonas vaginalis
Sci Rep, 8, 2018
|
|
5ZDA
 
 | |
5ZDC
 
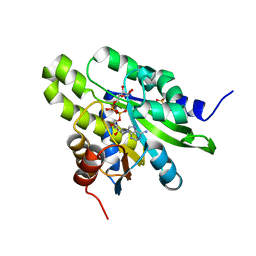 | | Crystal structure of poly(ADP-ribose) glycohydrolase (PARG) from Deinococcus radiodurans in complex with ADP-ribose (P32) | | Descriptor: | PHOSPHATE ION, [(2R,3S,4R,5R)-5-(6-AMINOPURIN-9-YL)-3,4-DIHYDROXY-OXOLAN-2-YL]METHYL [HYDROXY-[[(2R,3S,4R,5S)-3,4,5-TRIHYDROXYOXOLAN-2-YL]METHOXY]PHOSPHORYL] HYDROGEN PHOSPHATE, poly ADP-ribose glycohydrolase | | Authors: | Cho, C.C, Hsu, C.H. | | Deposit date: | 2018-02-23 | | Release date: | 2019-02-27 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.979 Å) | | Cite: | Structural and biochemical evidence supporting poly ADP-ribosylation in the bacterium Deinococcus radiodurans.
Nat Commun, 10, 2019
|
|
5ZDE
 
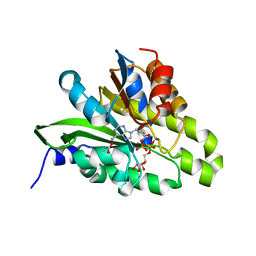 | | Crystal structure of poly(ADP-ribose) glycohydrolase (PARG) from Deinococcus radiodurans in complex with ADP-ribose (P3221) | | Descriptor: | Poly ADP-ribose glycohydrolase, [(2R,3S,4R,5R)-5-(6-AMINOPURIN-9-YL)-3,4-DIHYDROXY-OXOLAN-2-YL]METHYL[HYDROXY-[[(2R,3S,4R,5S)-3,4,5-TRIHYDROXYOXOLAN-2-YL]METHOXY]PHOSPHORYL] HYDROGEN PHOSPHATE | | Authors: | Cho, C.C, Hsu, C.H. | | Deposit date: | 2018-02-23 | | Release date: | 2019-02-27 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural and biochemical evidence supporting poly ADP-ribosylation in the bacterium Deinococcus radiodurans.
Nat Commun, 10, 2019
|
|
5ZDB
 
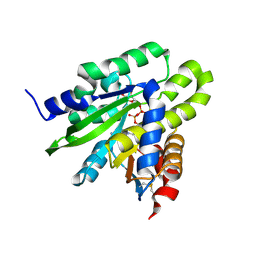 | | Crystal structure of poly(ADP-ribose) glycohydrolase (PARG) from Deinococcus radiodurans in complex with ADP-ribose (P21) | | Descriptor: | Poly ADP-ribose glycohydrolase, [(2R,3S,4R,5R)-5-(6-AMINOPURIN-9-YL)-3,4-DIHYDROXY-OXOLAN-2-YL]METHYL [HYDROXY-[[(2R,3S,4R,5S)-3,4,5-TRIHYDROXYOXOLAN-2-YL]METHOXY]PHOSPHORYL] HYDROGEN PHOSPHATE | | Authors: | Cho, C.C, Hsu, C.H. | | Deposit date: | 2018-02-23 | | Release date: | 2019-02-27 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.972 Å) | | Cite: | Structural and biochemical evidence supporting poly ADP-ribosylation in the bacterium Deinococcus radiodurans.
Nat Commun, 10, 2019
|
|
5ZDG
 
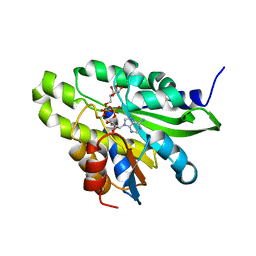 | | Crystal structure of poly(ADP-ribose) glycohydrolase (PARG) T267R mutant from Deinococcus radiodurans in complex with ADP-ribose | | Descriptor: | Poly APD-ribose glycohydrolase, [(2R,3S,4R,5R)-5-(6-AMINOPURIN-9-YL)-3,4-DIHYDROXY-OXOLAN-2-YL]METHYL[HYDROXY-[[(2R,3S,4R,5S)-3,4,5-TRIHYDROXYOXOLAN-2-YL]METHOXY]PHOSPHORYL] HYDROGEN PHOSPHATE | | Authors: | Cho, C.C, Hsu, C.H. | | Deposit date: | 2018-02-23 | | Release date: | 2019-02-27 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (2.594 Å) | | Cite: | Structural and biochemical evidence supporting poly ADP-ribosylation in the bacterium Deinococcus radiodurans.
Nat Commun, 10, 2019
|
|
5ZDD
 
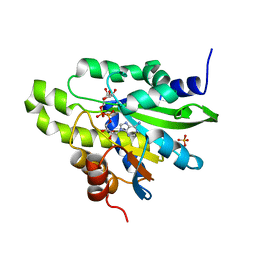 | | Crystal structure of poly(ADP-ribose) glycohydrolase (PARG) from Deinococcus radiodurans in complex with ADP-ribose (P212121) | | Descriptor: | PHOSPHATE ION, Poly ADP-ribose glycohydrolase, [(2R,3S,4R,5R)-5-(6-AMINOPURIN-9-YL)-3,4-DIHYDROXY-OXOLAN-2-YL]METHYL [HYDROXY-[[(2R,3S,4R,5S)-3,4,5-TRIHYDROXYOXOLAN-2-YL]METHOXY]PHOSPHORYL] HYDROGEN PHOSPHATE | | Authors: | Cho, C.C, Hsu, C.H. | | Deposit date: | 2018-02-23 | | Release date: | 2019-02-27 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.725 Å) | | Cite: | Structural and biochemical evidence supporting poly ADP-ribosylation in the bacterium Deinococcus radiodurans.
Nat Commun, 10, 2019
|
|
5ZDF
 
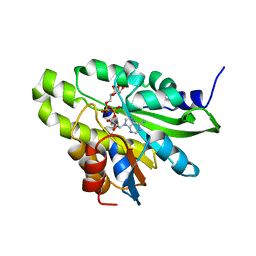 | | Crystal structure of poly(ADP-ribose) glycohydrolase (PARG) T267K mutant from Deinococcus radiodurans in complex with ADP-ribose | | Descriptor: | Poly ADP-ribose glycohydrolase, [(2R,3S,4R,5R)-5-(6-AMINOPURIN-9-YL)-3,4-DIHYDROXY-OXOLAN-2-YL]METHYL [HYDROXY-[[(2R,3S,4R,5S)-3,4,5-TRIHYDROXYOXOLAN-2-YL]METHOXY]PHOSPHORYL] HYDROGEN PHOSPHATE | | Authors: | Cho, C.C, Hsu, C.H. | | Deposit date: | 2018-02-23 | | Release date: | 2019-02-27 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.504 Å) | | Cite: | Structural and biochemical evidence supporting poly ADP-ribosylation in the bacterium Deinococcus radiodurans.
Nat Commun, 10, 2019
|
|
2MRD
 
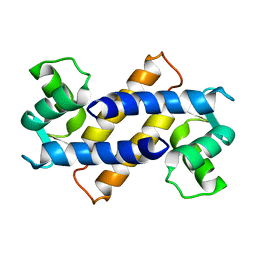 | |
1UD0
 
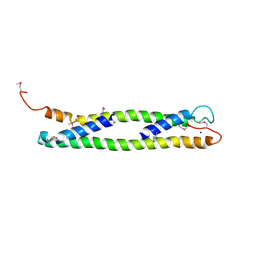 | | CRYSTAL STRUCTURE OF THE C-TERMINAL 10-kDA SUBDOMAIN OF HSC70 | | Descriptor: | 70 kDa heat-shock-like protein, SODIUM ION | | Authors: | Chou, C.C, Forouhar, F, Yeh, Y.H, Wang, C, Hsiao, C.D. | | Deposit date: | 2003-04-24 | | Release date: | 2004-05-11 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.45 Å) | | Cite: | Crystal structure of the C-terminal 10-kDa subdomain of Hsc70
J.BIOL.CHEM., 278, 2003
|
|
2GBZ
 
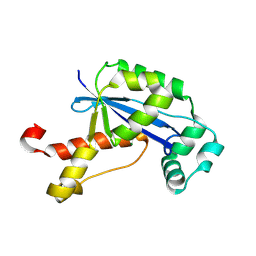 | | The Crystal Structure of XC847 from Xanthomonas campestris: a 3-5 Oligoribonuclease of DnaQ fold family with a Novel Opposingly-Shifted Helix | | Descriptor: | MAGNESIUM ION, Oligoribonuclease | | Authors: | Chin, K.H, Yang, C.Y, Chou, C.C, Wang, A.H.J, Chou, S.H. | | Deposit date: | 2006-03-12 | | Release date: | 2007-01-16 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The crystal structure of XC847 from Xanthomonas campestris: a 3'-5' oligoribonuclease of DnaQ fold family with a novel opposingly shifted helix
Proteins, 65, 2006
|
|
2FUJ
 
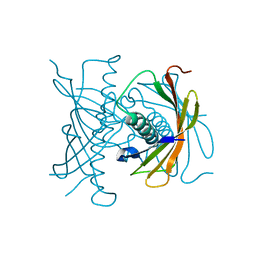 | |
2FA5
 
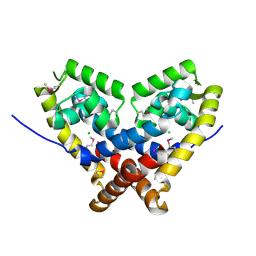 | | The crystal structure of an unliganded multiple antibiotic-resistance repressor (MarR) from Xanthomonas campestris | | Descriptor: | CHLORIDE ION, transcriptional regulator marR/emrR family | | Authors: | Chin, K.H, Tu, Z.L, Li, J.N, Chou, C.C, Wang, A.H.J, Chou, S.H. | | Deposit date: | 2005-12-06 | | Release date: | 2006-11-14 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The crystal structure of XC1739: a putative multiple antibiotic-resistance repressor (MarR) from Xanthomonas campestris at 1.8 A resolution
Proteins, 65, 2006
|
|
2FUK
 
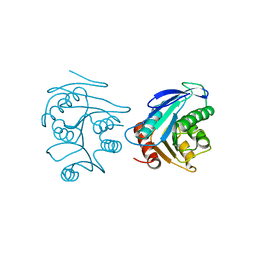 | | Crystal structure of XC6422 from Xanthomonas campestris: a member of a/b serine hydrolase without lid at 1.6 resolution | | Descriptor: | XC6422 protein | | Authors: | Yang, C.Y, Chin, K.H, Chou, C.C, Wang, A.H.J, Chou, S.H. | | Deposit date: | 2006-01-27 | | Release date: | 2006-07-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of XC6422 from Xanthomonas campestris at 1.6 A resolution: a small serine alpha/beta-hydrolase
Acta Crystallogr.,Sect.F, 62, 2006
|
|
2GU9
 
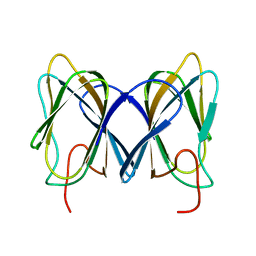 | |
2IO8
 
 | | E. coli Bifunctional glutathionylspermidine synthetase/amidase Incomplex with Mg2+ and ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Bifunctional glutathionylspermidine synthetase/amidase, CYSTEINE, ... | | Authors: | Pai, C.H, Chiang, B.Y, Ko, T.P, Chou, C.C, Chong, C.M, Yen, F.J, Coward, J.K, Wang, A.H.-J, Lin, C.H. | | Deposit date: | 2006-10-10 | | Release date: | 2006-12-12 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Dual binding sites for translocation catalysis by Escherichia coli glutathionylspermidine synthetase
Embo J., 25, 2006
|
|
2IOA
 
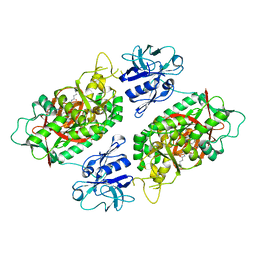 | | E. coli Bifunctional glutathionylspermidine synthetase/amidase Incomplex with Mg2+ and ADP and phosphinate inhibitor | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Bifunctional glutathionylspermidine synthetase/amidase, D-GAMMA-GLUTAMYL-N-{[(R)-{4-[(4-AMINOBUTYL)AMINO]BUTYL}(PHOSPHONOOXY)PHOSPHORYL]METHYL}-D-ALANINAMIDE, ... | | Authors: | Pai, C.H, Chiang, B.Y, Ko, T.P, Chou, C.C, Chong, C.M, Yen, F.J, Coward, J.K, Wang, A.H.-J, Lin, C.H. | | Deposit date: | 2006-10-10 | | Release date: | 2006-12-12 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Dual binding sites for translocation catalysis by Escherichia coli glutathionylspermidine synthetase
Embo J., 25, 2006
|
|
2IO9
 
 | | E. coli Bifunctional glutathionylspermidine synthetase/amidase Incomplex with Mg2+ ,GSH and ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Bifunctional glutathionylspermidine synthetase/amidase, GLUTATHIONE, ... | | Authors: | Pai, C.H, Chiang, B.Y, Ko, T.P, Chou, C.C, Chong, C.M, Yen, F.J, Coward, J.K, Wang, A.H.-J, Lin, C.H. | | Deposit date: | 2006-10-10 | | Release date: | 2006-12-12 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Dual binding sites for translocation catalysis by Escherichia coli glutathionylspermidine synthetase
Embo J., 25, 2006
|
|
2IOB
 
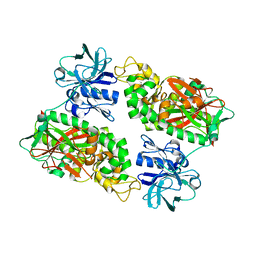 | | E. coli Bifunctional glutathionylspermidine synthetase/amidase Apo protein | | Descriptor: | Bifunctional glutathionylspermidine synthetase/amidase | | Authors: | Pai, C.H, Chiang, B.Y, Ko, T.P, Chou, C.C, Chong, C.M, Yen, F.J, Coward, J.K, Wang, A.H.-J, Lin, C.H. | | Deposit date: | 2006-10-10 | | Release date: | 2006-12-12 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Dual binding sites for translocation catalysis by Escherichia coli glutathionylspermidine synthetase
Embo J., 25, 2006
|
|
1H65
 
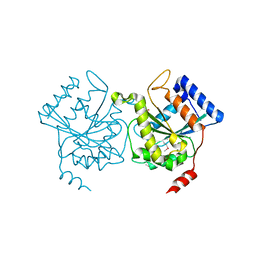 | | Crystal structure of pea Toc34 - a novel GTPase of the chloroplast protein translocon | | Descriptor: | CHLOROPLAST OUTER ENVELOPE PROTEIN OEP34, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION | | Authors: | Sun, Y.J, Forouhar, F, Li, H.M, Tu, S.L, Kao, S, Shr, H.L, Chou, C.C, Hsiao, C.D. | | Deposit date: | 2001-06-06 | | Release date: | 2002-01-29 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Pea Toc34 - a Novel Gtpase of the Chloroplast Protein Translocon
Nat.Struct.Biol., 9, 2002
|
|
1V4J
 
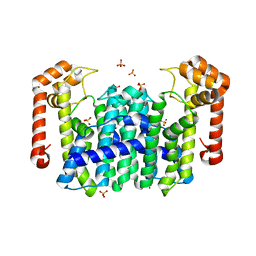 | | Crystal Structure of Octaprenyl Pyrophosphate Synthase from Hyperthermophilic Thermotoga maritima V73Y mutant | | Descriptor: | SULFATE ION, octoprenyl-diphosphate synthase | | Authors: | Guo, R.T, Kuo, C.J, Chou, C.C, Ko, T.P, Shr, H.L, Liang, P.H, Wang, A.H.-J. | | Deposit date: | 2003-11-14 | | Release date: | 2004-03-02 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Crystal Structure of Octaprenyl Pyrophosphate Synthase from Hyperthermophilic Thermotoga maritima and Mechanism of Product Chain Length Determination
J.Biol.Chem., 279, 2004
|
|
1VG7
 
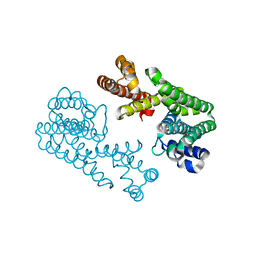 | | Crystal Structure Of Octaprenyl Pyrophosphate Synthase From Hyperthermophilic Thermotoga Maritima F132A/L128A/I123A/D62A mutant | | Descriptor: | octoprenyl-diphosphate synthase | | Authors: | Guo, R.T, Kuo, C.J, Ko, T.P, Chou, C.C, Liang, P.H, Wang, A.H.-J. | | Deposit date: | 2004-04-23 | | Release date: | 2004-05-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | A molecular ruler for chain elongation catalyzed by octaprenyl pyrophosphate synthase and its structure-based engineering to produce unprecedented long chain trans-prenyl products
Biochemistry, 43, 2004
|
|
1VG4
 
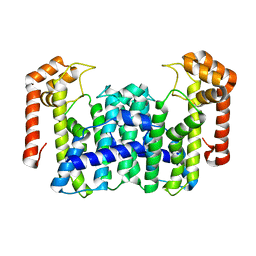 | | Crystal Structure Of Octaprenyl Pyrophosphate Synthase From Hyperthermophilic Thermotoga Maritima F132A/L128A mutant | | Descriptor: | octoprenyl-diphosphate synthase | | Authors: | Guo, R.T, Kuo, C.J, Ko, T.P, Chou, C.C, Liang, P.H, Wang, A.H.-J. | | Deposit date: | 2004-04-23 | | Release date: | 2004-05-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | A molecular ruler for chain elongation catalyzed by octaprenyl pyrophosphate synthase and its structure-based engineering to produce unprecedented long chain trans-prenyl products
Biochemistry, 43, 2004
|
|
1VG2
 
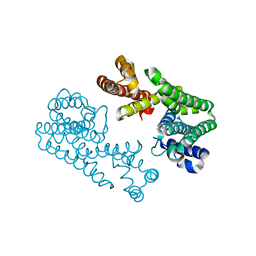 | | Crystal Structure Of Octaprenyl Pyrophosphate Synthase From Hyperthermophilic Thermotoga Maritima A76Y mutant | | Descriptor: | octoprenyl-diphosphate synthase | | Authors: | Guo, R.T, Kuo, C.J, Ko, T.P, Chou, C.C, Liang, P.H, Wang, A.H.-J. | | Deposit date: | 2004-04-23 | | Release date: | 2004-05-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | A molecular ruler for chain elongation catalyzed by octaprenyl pyrophosphate synthase and its structure-based engineering to produce unprecedented long chain trans-prenyl products
Biochemistry, 43, 2004
|
|
