7D83
 
 | | Crystal structure of HIV-1 integrase catalytic core domain in complex with 2-(tert-butoxy)-2-(2-(3-cyclohexylureido)-3,6-dimethyl-5-(5-methylchroman-6-yl)pyridin-4-yl)acetic acid | | Descriptor: | (2S)-2-[2-(cyclohexylcarbamoylamino)-3,6-dimethyl-5-(5-methyl-3,4-dihydro-2H-chromen-6-yl)pyridin-4-yl]-2-[(2-methylpropan-2-yl)oxy]ethanoic acid, Integrase, SULFATE ION | | Authors: | Sugiyama, S, Sekiguchi, Y. | | Deposit date: | 2020-10-07 | | Release date: | 2021-01-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Discovery of novel HIV-1 integrase-LEDGF/p75 allosteric inhibitors based on a pyridine scaffold forming an intramolecular hydrogen bond.
Bioorg.Med.Chem.Lett., 33, 2020
|
|
6JKI
 
 | | Crystal structure and catalytic mechanism of the essential m1G37 tRNA methyltransferase TrmD from Pseudomonas aeruginosa | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, MANGANESE (II) ION, ... | | Authors: | Jaroensuk, J, Liew, C.W, Atichartpongkul, S, Chionh, Y.H, Wong, Y.H, Zhong, W.H, McBee, M.E, Thongdee, N, Prestwich, E.G, DeMott, M.S, Mongkolsuk, S, Dedon, P.C, Lescar, J, Fuangthong, M. | | Deposit date: | 2019-03-01 | | Release date: | 2019-07-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Crystal structure and catalytic mechanism of the essential m1G37 tRNA methyltransferase TrmD fromPseudomonas aeruginosa.
Rna, 25, 2019
|
|
1V9T
 
 | | Structure of E. coli cyclophilin B K163T mutant bound to succinyl-ALA-PRO-ALA-P-nitroanilide | | Descriptor: | (SIN)APA(NIT), cyclophilin B | | Authors: | Konno, M, Sano, Y, Okudaira, K, Kawaguchi, Y, Yamagishi-Ohmori, Y, Fushinobu, S, Matsuzawa, H. | | Deposit date: | 2004-02-03 | | Release date: | 2004-09-21 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Escherichia coli cyclophilin B binds a highly distorted form of trans-prolyl peptide isomer
Eur.J.Biochem., 271, 2004
|
|
3X39
 
 | | Domain-swapped dimer of Pseudomonas aeruginosa cytochrome c551 | | Descriptor: | Cytochrome c-551, HEME C | | Authors: | Nagao, S, Ueda, M, Osuka, H, Komori, H, Kamikubo, H, Kataoka, M, Higuchi, Y, Hirota, S. | | Deposit date: | 2015-01-16 | | Release date: | 2015-04-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Domain-Swapped Dimer of Pseudomonas aeruginosa Cytochrome c551: Structural Insights into Domain Swapping of Cytochrome c Family Proteins
Plos One, 10, 2015
|
|
4H4S
 
 | | Crystal Structure of Ferredoxin reductase, BphA4 E175C/Q177G mutant (reduced form) | | Descriptor: | Biphenyl dioxygenase ferredoxin reductase subunit, FLAVIN-ADENINE DINUCLEOTIDE, FORMIC ACID, ... | | Authors: | Nishizawa, A, Harada, A, Senda, M, Tachihara, Y, Muramatsu, D, Kishigami, S, Mori, S, Sugiyama, K, Senda, T, Kimura, S. | | Deposit date: | 2012-09-18 | | Release date: | 2013-10-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Random Mutagenesis with the Project Assessment for Complete Conversion of Co-Factor Specificity of a Ferredoxin Reductase BphA4
To be Published
|
|
4H4W
 
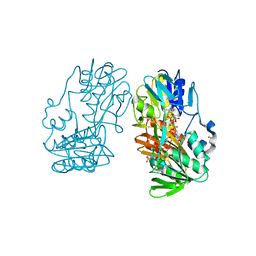 | | Crystal Structure of Ferredoxin reductase, BphA4 E175C/T176R/Q177G mutant (reduced form) | | Descriptor: | Biphenyl dioxygenase ferredoxin reductase subunit, FLAVIN-ADENINE DINUCLEOTIDE, FORMIC ACID, ... | | Authors: | Nishizawa, A, Harada, A, Senda, M, Tachihara, Y, Muramatsu, D, Kishigami, S, Mori, S, Sugiyama, K, Senda, T, Kimura, S. | | Deposit date: | 2012-09-18 | | Release date: | 2013-10-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Random Mutagenesis with the Project Assessment for Complete Conversion of Co-Factor Specificity of a Ferredoxin Reductase BphA4
To be Published
|
|
4H4U
 
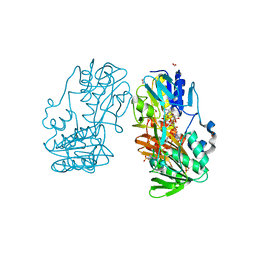 | | Crystal Structure of Ferredoxin reductase, BphA4 T176R mutant (reduced form) | | Descriptor: | Biphenyl dioxygenase ferredoxin reductase subunit, FLAVIN-ADENINE DINUCLEOTIDE, FORMIC ACID, ... | | Authors: | Nishizawa, A, Harada, A, Senda, M, Tachihara, Y, Muramatsu, D, Kishigami, S, Mori, S, Sugiyama, K, Senda, T, Kimura, S. | | Deposit date: | 2012-09-18 | | Release date: | 2013-10-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Random Mutagenesis with the Project Assessment for Complete Conversion of Co-Factor Specificity of a Ferredoxin Reductase BphA4
To be Published
|
|
7WCE
 
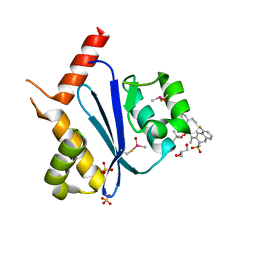 | | Crystal structure of HIV-1 integrase catalytic core domain in complex with (2S)-2-(tert-Butoxy)-2-(10-fluoro-2-(2-hydroxy-4-methylphenyl)-1,4-dimethyl-5-(methylsulfonyl)-5,6-dihydrophenanthridin-3-yl)acetic acid | | Descriptor: | (2S)-2-[10-fluoranyl-1,4-dimethyl-2-(4-methyl-2-oxidanyl-phenyl)-5-methylsulfonyl-6H-phenanthridin-3-yl]-2-[(2-methylpropan-2-yl)oxy]ethanoic acid, GLYCEROL, Integrase catalytic, ... | | Authors: | Taoda, Y, Sekiguchi, Y. | | Deposit date: | 2021-12-20 | | Release date: | 2022-09-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Discovery of tricyclic HIV-1 integrase-LEDGF/p75 allosteric inhibitors by intramolecular direct arylation reaction.
Bioorg.Med.Chem.Lett., 64, 2022
|
|
7WMC
 
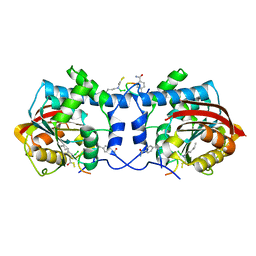 | | Crystal structure of macrocyclic peptide 1 bound to human Nicotinamide N-methyltransferase | | Descriptor: | Nicotinamide N-methyltransferase, Peptide1 | | Authors: | Yoshida, S, Uehara, S, Kondo, N, Takahashi, Y, Yamamoto, S, Kameda, A, Kawagoe, S, Inoue, N, Yamada, M, Yoshimura, N, Tachibana, Y. | | Deposit date: | 2022-01-14 | | Release date: | 2022-08-31 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Peptide-to-Small Molecule: A Pharmacophore-Guided Small Molecule Lead Generation Strategy from High-Affinity Macrocyclic Peptides.
J.Med.Chem., 65, 2022
|
|
7WMT
 
 | | Crystal structure of small molecule 13 bound to human Nicotinamide N-methyltransferase | | Descriptor: | Nicotinamide N-methyltransferase, [(2~{R},4~{S})-4-[2-(aminomethyl)imidazol-1-yl]-2-[1-[(4-chlorophenyl)methyl]-5-methyl-indol-2-yl]pyrrolidin-1-yl]-(1~{H}-pyrrolo[2,3-b]pyridin-5-yl)methanone | | Authors: | Yoshida, S, Uehara, S, Kondo, N, Takahashi, Y, Yamamoto, S, Kameda, A, Kawagoe, S, Inoue, N, Yamada, M, Yoshimura, N, Tachibana, Y. | | Deposit date: | 2022-01-17 | | Release date: | 2022-08-31 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Peptide-to-Small Molecule: A Pharmacophore-Guided Small Molecule Lead Generation Strategy from High-Affinity Macrocyclic Peptides.
J.Med.Chem., 65, 2022
|
|
1V9F
 
 | | Crystal structure of catalytic domain of pseudouridine synthase RluD from Escherichia coli | | Descriptor: | PHOSPHATE ION, Ribosomal large subunit pseudouridine synthase D | | Authors: | Mizutani, K, Machida, Y, Unzai, S, Park, S.-Y, Tame, J.R.H. | | Deposit date: | 2004-01-26 | | Release date: | 2004-05-18 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structures of the catalytic domains of pseudouridine synthases RluC and RluD from Escherichia coli
Biochemistry, 43, 2004
|
|
1GIR
 
 | | CRYSTAL STRUCTURE OF THE ENZYMATIC COMPONET OF IOTA-TOXIN FROM CLOSTRIDIUM PERFRINGENS WITH NADPH | | Descriptor: | IOTA TOXIN COMPONENT IA, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Tsuge, H, Nagahama, M, Nishimura, H, Hisatsune, J, Sakaguchi, Y, Itogawa, Y, Katunuma, N, Sakurai, J. | | Deposit date: | 2001-03-12 | | Release date: | 2003-01-14 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure and Site-directed Mutagenesis of Enzymatic Components from Clostridium perfringens Iota-toxin
J.MOL.BIOL., 325, 2003
|
|
1VAI
 
 | | Structure of e. coli cyclophilin B K163T mutant bound to n-acetyl-ala-ala-pro-ala-7-amino-4-methylcoumarin | | Descriptor: | (ACE)AAPA(MCM), cyclophilin B | | Authors: | Konno, M, Sano, Y, Okudaira, K, Kawaguchi, Y, Yamagishi-Ohmori, Y, Fushinobu, S, Matsuzawa, H. | | Deposit date: | 2004-02-17 | | Release date: | 2004-09-21 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Escherichia coli cyclophilin B binds a highly distorted form of trans-prolyl peptide isomer
Eur.J.Biochem., 271, 2004
|
|
1J0O
 
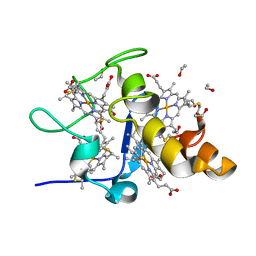 | | High Resolution Crystal Structure of the wild type Tetraheme Cytochrome c3 from Desulfovibrio vulgaris Miyazaki F | | Descriptor: | Cytochrome c3, ETHANOL, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Ozawa, K, Yasukawa, F, Kumagai, J, Ohmura, T, Cusanvich, M.A, Tomimoto, Y, Ogata, H, Higuchi, Y, Akutsu, H. | | Deposit date: | 2002-11-19 | | Release date: | 2003-11-19 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Role of the aromatic ring of Tyr43 in tetraheme cytochrome c(3) from Desulfovibrio vulgaris Miyazaki F.
Biophys.J., 85, 2003
|
|
1J2A
 
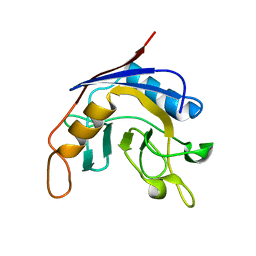 | | Structure of E. coli cyclophilin B K163T mutant | | Descriptor: | cyclophilin B | | Authors: | Konno, M, Sano, Y, Okudaira, K, Kawaguchi, Y, Yamagishi-Ohmori, Y, Fushinobu, S, Matsuzawa, H. | | Deposit date: | 2002-12-26 | | Release date: | 2004-02-10 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Escherichia coli cyclophilin B binds a highly distorted form of trans-prolyl peptide isomer
Eur.J.Biochem., 271, 2004
|
|
4H4T
 
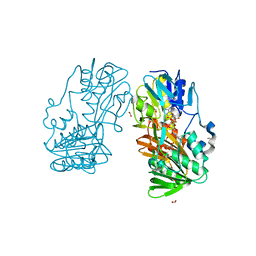 | | Crystal Structure of Ferredoxin reductase, BphA4 T176R mutant (oxidized form) | | Descriptor: | Biphenyl dioxygenase ferredoxin reductase subunit, FLAVIN-ADENINE DINUCLEOTIDE, FORMIC ACID, ... | | Authors: | Nishizawa, A, Harada, A, Senda, M, Tachihara, Y, Muramatsu, D, Kishigami, S, Mori, S, Sugiyama, K, Senda, T, Kimura, S. | | Deposit date: | 2012-09-18 | | Release date: | 2013-10-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Random Mutagenesis with the Project Assessment for Complete Conversion of Co-Factor Specificity of a Ferredoxin Reductase BphA4
To be Published
|
|
4H4R
 
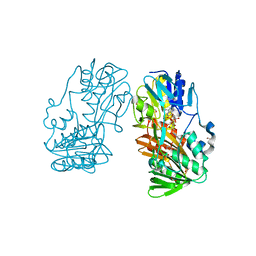 | | Crystal Structure of Ferredoxin reductase, BphA4 E175C/Q177G mutant (oxidized form) | | Descriptor: | Biphenyl dioxygenase ferredoxin reductase subunit, FLAVIN-ADENINE DINUCLEOTIDE, FORMIC ACID, ... | | Authors: | Nishizawa, A, Harada, A, Senda, M, Tachihara, Y, Muramatsu, D, Kishigami, S, Mori, S, Sugiyama, K, Senda, T, Kimura, S. | | Deposit date: | 2012-09-18 | | Release date: | 2013-10-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Random Mutagenesis with the Project Assessment for Complete Conversion of Co-Factor Specificity of a Ferredoxin Reductase BphA4
To be Published
|
|
1VGN
 
 | | Structure-based design of the irreversible inhibitors to metallo--lactamase (IMP-1) | | Descriptor: | 3-OXO-3-[(3-OXOPROPYL)SULFANYL]PROPANE-1-THIOLATE, ACETIC ACID, Beta-lactamase IMP-1, ... | | Authors: | Kurosaki, H, Yamaguchi, Y, Higashi, T, Soga, K, Matsueda, S, Misumi, S, Yamagata, Y, Arakawa, Y, Goto, M. | | Deposit date: | 2004-04-27 | | Release date: | 2005-06-21 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.63 Å) | | Cite: | Irreversible Inhibition of Metallo-beta-lactamase (IMP-1) by 3-(3-Mercaptopropionylsulfanyl)propionic Acid Pentafluorophenyl Ester
Angew.Chem.Int.Ed.Engl., 44, 2005
|
|
3UX0
 
 | | Crystal structure of human 14-3-3 sigma in complex with TASK-3 peptide and stabilizer Fusicoccin H | | Descriptor: | (4R,5R,6R,6aS,9S,9aE,10aR)-5-hydroxy-9-(hydroxymethyl)-6,10a-dimethyl-3-(propan-2-yl)-1,2,4,5,6,6a,7,8,9,10a-decahydrodicyclopenta[a,d][8]annulen-4-yl alpha-D-gulopyranoside, 14-3-3 protein sigma, CHLORIDE ION, ... | | Authors: | Thiel, P, Bartel, M, Anders, C, Higuchi, Y, Schumacher, B, Kato, N, Ottmann, C. | | Deposit date: | 2011-12-03 | | Release date: | 2013-01-02 | | Last modified: | 2013-05-01 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | A semisynthetic fusicoccane stabilizes a protein-protein interaction and enhances the expression of k(+) channels at the cell surface.
Chem.Biol., 20, 2013
|
|
1WMN
 
 | | Crystal structure of topaquinone-containing amine oxidase activated by cobalt ion | | Descriptor: | COBALT (II) ION, Phenylethylamine oxidase | | Authors: | Okajima, T, Kishishita, S, Chiu, Y.C, Murakawa, T, Kim, M, Yamaguchi, H, Hirota, S, Kuroda, S, Tanizawa, K. | | Deposit date: | 2004-07-13 | | Release date: | 2005-08-02 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Reinvestigation of metal ion specificity for quinone cofactor biogenesis in bacterial copper amine oxidase
Biochemistry, 44, 2005
|
|
1WMO
 
 | | Crystal structure of topaquinone-containing amine oxidase activated by nickel ion | | Descriptor: | NICKEL (II) ION, Phenylethylamine oxidase | | Authors: | Okajima, T, Kishishita, S, Chiu, Y.C, Murakawa, T, Kim, M, Yamaguchi, H, Hirota, S, Kuroda, S, Tanizawa, K. | | Deposit date: | 2004-07-13 | | Release date: | 2005-08-02 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Reinvestigation of metal ion specificity for quinone cofactor biogenesis in bacterial copper amine oxidase
Biochemistry, 44, 2005
|
|
4P7W
 
 | | L-proline-bound L-proline cis-4-hydroxylase | | Descriptor: | 2-OXOGLUTARIC ACID, COBALT (II) ION, L-proline cis-4-hydroxylase, ... | | Authors: | Shomura, Y, Koketsu, K, Moriwaki, K, Hayashi, M, Mitsuhashi, S, Hara, R, Kino, K, Higuchi, Y. | | Deposit date: | 2014-03-28 | | Release date: | 2014-09-17 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Refined Regio- and Stereoselective Hydroxylation of l-Pipecolic Acid by Protein Engineering of l-Proline cis-4-Hydroxylase Based on the X-ray Crystal Structure.
Acs Synth Biol, 4, 2015
|
|
6IQX
 
 | | High resolution structure of bilirubin oxidase from Myrothecium verrucaria - M467Q mutant, aerobically prepared | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, Bilirubin oxidase, ... | | Authors: | Shibata, N, Akter, M, Higuchi, Y. | | Deposit date: | 2018-11-09 | | Release date: | 2018-11-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.432 Å) | | Cite: | Redox Potential-Dependent Formation of an Unusual His-Trp Bond in Bilirubin Oxidase.
Chemistry, 24, 2018
|
|
7V5P
 
 | | The dimeric structure of G80A/H81A myoglobin | | Descriptor: | Myoglobin, OXYGEN ATOM, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Xie, C, Nagao, S, Shibata, N, Higuchi, Y, Hirota, S. | | Deposit date: | 2021-08-17 | | Release date: | 2022-06-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.16 Å) | | Cite: | Experimental and theoretical study on converting myoglobin into a stable domain-swapped dimer by utilizing a tight hydrogen bond network at the hinge region.
Rsc Adv, 11, 2021
|
|
6IQZ
 
 | | High resolution structure of bilirubin oxidase from Myrothecium verrucaria - wild type | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Bilirubin oxidase, COPPER (II) ION, ... | | Authors: | Shibata, N, Akter, M, Higuchi, Y. | | Deposit date: | 2018-11-09 | | Release date: | 2018-11-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Redox Potential-Dependent Formation of an Unusual His-Trp Bond in Bilirubin Oxidase.
Chemistry, 24, 2018
|
|
