5DUI
 
 | |
2H8R
 
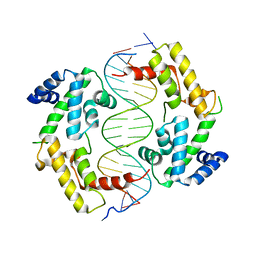 | | Hepatocyte Nuclear Factor 1b bound to DNA: MODY5 Gene Product | | Descriptor: | 5'-D(*CP*TP*TP*GP*GP*TP*TP*AP*AP*TP*AP*AP*TP*TP*CP*AP*CP*CP*AP*G)-3', 5'-D(*GP*CP*TP*GP*GP*TP*GP*AP*AP*TP*TP*AP*TP*TP*AP*AP*CP*CP*AP*A)-3', Hepatocyte nuclear factor 1-beta | | Authors: | Lu, P, Rha, G.B, Chi, Y.I. | | Deposit date: | 2006-06-07 | | Release date: | 2007-06-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural basis of disease-causing mutations in hepatocyte nuclear factor 1beta.
Biochemistry, 46, 2007
|
|
1Q2H
 
 | | Phenylalanine Zipper Mediates APS Dimerization | | Descriptor: | adaptor protein with pleckstrin homology and src homology 2 domains | | Authors: | Dhe-Paganon, S, Werner, E.D, Nishi, M, Chi, Y.-I, Shoelson, S.E. | | Deposit date: | 2003-07-24 | | Release date: | 2004-08-03 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A phenylalanine zipper mediates APS dimerization.
Nat.Struct.Mol.Biol., 11, 2004
|
|
1PZL
 
 | | Crystal structure of HNF4a LBD in complex with the ligand and the coactivator SRC-1 peptide | | Descriptor: | Hepatocyte nuclear factor 4-alpha, MYRISTIC ACID, steroid receptor coactivator-1 | | Authors: | Duda, K, Chi, Y.-I, Dhe-paganon, S, Shoelson, S. | | Deposit date: | 2003-07-11 | | Release date: | 2004-06-01 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Basis for HNF-4alpha Activation by Ligand and Coactivator Binding
J.Biol.Chem., 279, 2004
|
|
3KSI
 
 | |
3KSH
 
 | |
3KE4
 
 | |
3KE5
 
 | |
3KSF
 
 | |
3KSG
 
 | |
4KZ2
 
 | | Crystal Structure of phi29 pRNA 3WJ Core | | Descriptor: | MANGANESE (II) ION, phi29 pRNA 3WJ core RNA 16 mer, phi29 pRNA 3WJ core RNA 18 mer, ... | | Authors: | Zhang, H, Endrizzi, J.A, Shu, Y, Haque, F, Sauter, C, Guo, P, Chi, Y.-I. | | Deposit date: | 2013-05-29 | | Release date: | 2013-10-16 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Crystal structure of 3WJ core revealing divalent ion-promoted thermostability and assembly of the Phi29 hexameric motor pRNA.
Rna, 19, 2013
|
|
3H1J
 
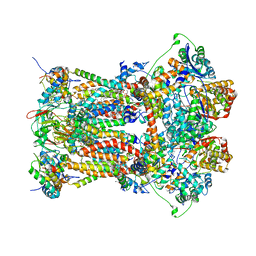 | | Stigmatellin-bound cytochrome bc1 complex from chicken | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, CARDIOLIPIN, Coenzyme Q10, ... | | Authors: | Zhang, Z, Huang, L, Shulmeister, V.M, Chi, Y.-I, Kim, K.K, Hung, L.-W, Crofts, A.R, Berry, E.A, Kim, S.-H. | | Deposit date: | 2009-04-12 | | Release date: | 2009-04-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Electron Transfer by Domain Movement in Cytochrome Bc1
Nature, 392, 1998
|
|
3FS1
 
 | |
4NAR
 
 | | Crystal Structure of the Q9WYS3 protein from Thermotoga maritima. Northeast Structural Genomics Consortium Target VR152 | | Descriptor: | ACETATE ION, Putative uronate isomerase, SULFATE ION | | Authors: | Vorobiev, S, Lew, S, Seetharaman, J, Chi, Y, Xiao, R, Maglaqui, M, Lee, D, Everett, J.K, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2013-10-22 | | Release date: | 2013-11-06 | | Method: | X-RAY DIFFRACTION (2.388 Å) | | Cite: | Crystal Structure of the Q9WYS3 protein from Thermotoga maritima.
To be Published
|
|
3DRE
 
 | |
3DRB
 
 | | Crystal structure of Human Brain-type Creatine Kinase | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Creatine kinase B-type, MAGNESIUM ION | | Authors: | Moon, J.H, Bong, S.M, Hwang, K.Y, Chi, Y.M. | | Deposit date: | 2008-07-11 | | Release date: | 2009-03-17 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural studies of human brain-type creatine kinase complexed with the ADP-Mg2+-NO3- -creatine transition-state analogue complex
Febs Lett., 582, 2008
|
|
402D
 
 | | 5'-R(*CP*GP*CP*CP*AP*GP*CP*G)-3' | | Descriptor: | RNA(5'-R(*CP*GP*CP*CP*AP*GP*CP*G)-3') | | Authors: | Jang, S.B, Hung, L.-W, Chi, Y.-I, Holbrook, E.L, Carter, R, Holbrook, S.R. | | Deposit date: | 1998-06-04 | | Release date: | 1998-06-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of an RNA internal loop consisting of tandem C-A+ base pairs.
Biochemistry, 37, 1998
|
|
3H1I
 
 | | Stigmatellin and antimycin bound cytochrome bc1 complex from chicken | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, 2-METHYL-BUTYRIC ACID 3-(3-FORMYLAMINO-2-HYDROXY-BENZOYLAMINO)-8-HEPTYL-2,6-DIMETHYL-4,9-DIOXO-[1,5]DIOXONAN-7-YL ESTER, CARDIOLIPIN, ... | | Authors: | Zhang, Z, Huang, L, Shulmeister, V.M, Chi, Y.I, Kim, K.K, Hung, L.W, Crofts, A.R, Berry, E.A, Kim, S.H. | | Deposit date: | 2009-04-12 | | Release date: | 2009-04-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.53 Å) | | Cite: | Electron Transfer by Domain Movement in Cytochrome Bc1
Nature, 392, 1998
|
|
3H1H
 
 | | Cytochrome bc1 complex from chicken | | Descriptor: | 1,2-Dioleoyl-sn-glycero-3-phosphoethanolamine, CARDIOLIPIN, CYTOCHROME C1, ... | | Authors: | Zhang, Z, Huang, L, Shulmeister, V.M, Chi, Y.I, Kim, K.K, Hung, L.W, Crofts, A.R, Berry, E.A, Kim, S.H. | | Deposit date: | 2009-04-12 | | Release date: | 2009-04-28 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.16 Å) | | Cite: | Electron Transfer by Domain Movement in Cytochrome Bc1
Nature, 392, 1998
|
|
4HYE
 
 | |
3UYU
 
 | | Structural basis for the antifreeze activity of an ice-binding protein (LeIBP) from Arctic yeast | | Descriptor: | Antifreeze protein, GLYCEROL | | Authors: | Lee, J.H, Park, A.K, Do, H, Park, K.S, Moh, S.H, Chi, Y.M, Kim, H.J. | | Deposit date: | 2011-12-06 | | Release date: | 2012-02-15 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Structural basis for the antifreeze activity of an ice-binding protein from an Arctic yeast.
J.Biol.Chem., 2012
|
|
1BCC
 
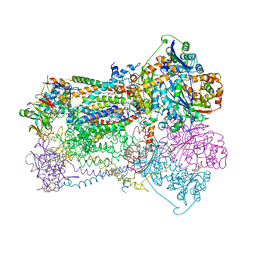 | | CYTOCHROME BC1 COMPLEX FROM CHICKEN | | Descriptor: | 1,2-Dioleoyl-sn-glycero-3-phosphoethanolamine, FE2/S2 (INORGANIC) CLUSTER, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Zhang, Z, Huang, L, Shulmeister, V.M, Chi, Y.-I, Kim, K.K, Hung, L.-W, Crofts, A.R, Berry, E.A, Kim, S.-H. | | Deposit date: | 1998-03-23 | | Release date: | 1998-08-19 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.16 Å) | | Cite: | Electron transfer by domain movement in cytochrome bc1.
Nature, 392, 1998
|
|
4OHT
 
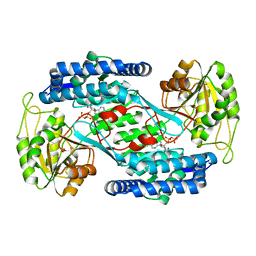 | | Crystal structure of succinic semialdehyde dehydrogenase from Streptococcus pyogenes in complex with NADP+ as the cofactor | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Succinate-semialdehyde dehydrogenase | | Authors: | Park, S.A, Jang, E.H, Chi, Y.M, Lee, K.S. | | Deposit date: | 2014-01-18 | | Release date: | 2014-12-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Kinetic and Structural Characterization for Cofactor Preference of Succinic Semialdehyde Dehydrogenase from Streptococcus pyogenes.
Mol.Cells, 37, 2014
|
|
4OGD
 
 | |
3QAW
 
 | |
