3WCR
 
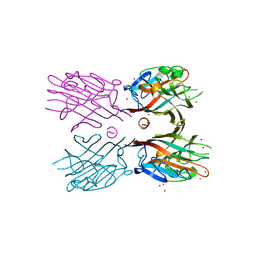 | | Crystal structure of plant lectin (ligand-free form) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, BROMIDE ION, Erythroagglutinin | | 著者 | Nagae, M, Yamaguchi, Y. | | 登録日 | 2013-05-31 | | 公開日 | 2014-04-09 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (2.45 Å) | | 主引用文献 | Phytohemagglutinin from Phaseolus vulgaris (PHA-E) displays a novel glycan recognition mode using a common legume lectin fold
Glycobiology, 24, 2014
|
|
3WBP
 
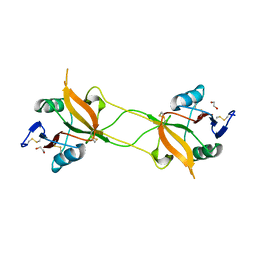 | | Crystal structure of carbohydrate recognition domain of Blood Dendritic Cell Antigen-2 (BDCA2) lectin (crystal form-1) | | 分子名称: | 1,2-ETHANEDIOL, C-type lectin domain family 4 member C | | 著者 | Nagae, M, Ikeda, A, Kitago, Y, Matsumoto, N, Yamamoto, K, Yamaguchi, Y. | | 登録日 | 2013-05-20 | | 公開日 | 2013-12-25 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Crystal structures of carbohydrate recognition domain of blood dendritic cell antigen-2 (BDCA2) reveal a common domain-swapped dimer.
Proteins, 82, 2014
|
|
3WWK
 
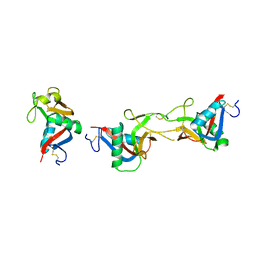 | | Crystal structure of CLEC-2 in complex with rhodocytin | | 分子名称: | C-type lectin domain family 1 member B, Snaclec rhodocytin subunit alpha, Snaclec rhodocytin subunit beta | | 著者 | Nagae, M, Morita-Matsumoto, K, Kato, M, Kato-Kaneko, M, Kato, Y, Yamaguchi, Y. | | 登録日 | 2014-06-20 | | 公開日 | 2014-10-22 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.98 Å) | | 主引用文献 | A Platform of C-type Lectin-like Receptor CLEC-2 for Binding O-Glycosylated Podoplanin and Nonglycosylated Rhodocytin
Structure, 22, 2014
|
|
3WBR
 
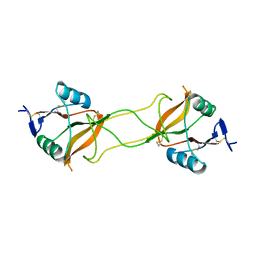 | | Crystal structure of carbohydrate recognition domain of Blood Dendritic Cell Antigen-2 (BDCA2) lectin (crystal form-3) | | 分子名称: | C-type lectin domain family 4 member C | | 著者 | Nagae, M, Ikeda, A, Kitago, Y, Matsumoto, N, Yamamoto, K, Yamaguchi, Y. | | 登録日 | 2013-05-20 | | 公開日 | 2013-12-25 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Crystal structures of carbohydrate recognition domain of blood dendritic cell antigen-2 (BDCA2) reveal a common domain-swapped dimer.
Proteins, 82, 2014
|
|
3WSR
 
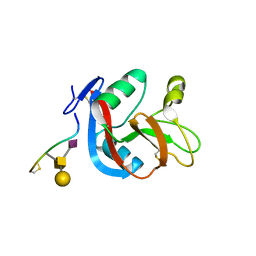 | | Crystal structure of CLEC-2 in complex with O-glycosylated podoplanin | | 分子名称: | C-type lectin domain family 1 member B, Peptide from Podoplanin, beta-D-galactopyranose-(1-3)-[N-acetyl-alpha-neuraminic acid-(2-6)]2-acetamido-2-deoxy-alpha-D-galactopyranose | | 著者 | Nagae, M, Morita-Matsumoto, K, Kato, M, Kato-Kaneko, M, Kato, Y, Yamaguchi, Y. | | 登録日 | 2014-03-20 | | 公開日 | 2014-10-22 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.91 Å) | | 主引用文献 | A Platform of C-type Lectin-like Receptor CLEC-2 for Binding O-Glycosylated Podoplanin and Nonglycosylated Rhodocytin
Structure, 22, 2014
|
|
3J6P
 
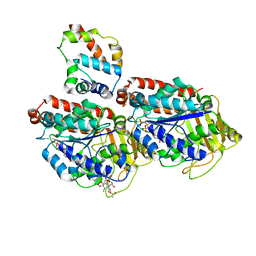 | | Pseudo-atomic model of dynein microtubule binding domain-tubulin complex based on a cryoEM map | | 分子名称: | Dynein heavy chain, cytoplasmic, GUANOSINE-5'-DIPHOSPHATE, ... | | 著者 | Uchimura, S, Fujii, T, Takazaki, H, Ayukawa, R, Nishikawa, Y, Minoura, I, Hachikubo, Y, Kurisu, G, Sutoh, K, Kon, T, Namba, K, Muto, E. | | 登録日 | 2014-03-20 | | 公開日 | 2014-12-31 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (8.2 Å) | | 主引用文献 | A flipped ion pair at the dynein-microtubule interface is critical for dynein motility and ATPase activation
J.Cell Biol., 208, 2015
|
|
4YSN
 
 | | Structure of aminoacid racemase in complex with PLP | | 分子名称: | PYRIDOXAL-5'-PHOSPHATE, Putative 4-aminobutyrate aminotransferase | | 著者 | Sakuraba, H, Mutaguchi, Y, Hayashi, J, Ohshima, T. | | 登録日 | 2015-03-17 | | 公開日 | 2016-04-20 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.94 Å) | | 主引用文献 | Crystal structure of the novel amino-acid racemase isoleucine 2-epimerase from Lactobacillus buchneri.
Acta Crystallogr D Struct Biol, 73, 2017
|
|
4WR5
 
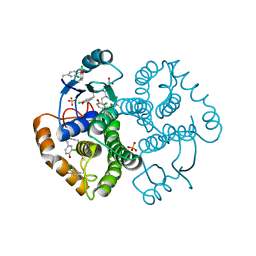 | | Crystal Structure of GST Mutated with Halogenated Tyrosine (7cGST-1) | | 分子名称: | GLUTATHIONE, Glutathione S-transferase class-mu 26 kDa isozyme, SULFATE ION | | 著者 | Akasaka, R, Kawazoe, M, Tomabechi, Y, Ohtake, K, Itagaki, T, Takemoto, C, Shirouzu, M, Yokoyama, S, Sakamoto, K. | | 登録日 | 2014-10-23 | | 公開日 | 2015-08-19 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.93 Å) | | 主引用文献 | Protein stabilization utilizing a redefined codon
Sci Rep, 5, 2015
|
|
4WR4
 
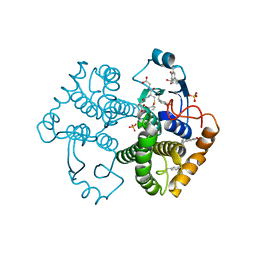 | | Crystal Structure of GST Mutated with Halogenated Tyrosine (7bGST-1) | | 分子名称: | GLUTATHIONE, Glutathione S-transferase class-mu 26 kDa isozyme, SULFATE ION | | 著者 | Akasaka, R, Kawazoe, M, Tomabechi, Y, Ohtake, K, Itagaki, T, Takemoto, C, Shirouzu, M, Yokoyama, S, Sakamoto, K. | | 登録日 | 2014-10-23 | | 公開日 | 2015-08-19 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Protein stabilization utilizing a redefined codon
Sci Rep, 5, 2015
|
|
7PN0
 
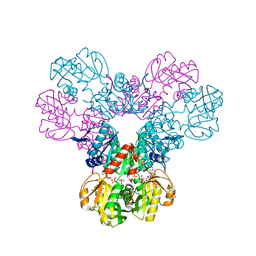 | | Crystal structure of the Phosphorybosylpyrophosphate synthetase II from Thermus thermophilus at R32 space group | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, Ribose-phosphate pyrophosphokinase, SULFATE ION | | 著者 | Timofeev, V.I, Abramchik, Y.A, Kostromina, M.A, Esipov, R.S, Kuranova, I.P. | | 登録日 | 2021-09-04 | | 公開日 | 2021-09-29 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Crystal structure of the Phosphorybosylpyrophosphate synthetase II from Thermus thermophilus at R32 space group
To Be Published
|
|
3WOG
 
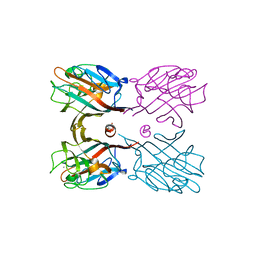 | | Crystal structure plant lectin in complex with ligand | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose, CALCIUM ION, ... | | 著者 | Nagae, M, Yamaguchi, Y. | | 登録日 | 2013-12-26 | | 公開日 | 2014-04-09 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Phytohemagglutinin from Phaseolus vulgaris (PHA-E) displays a novel glycan recognition mode using a common legume lectin fold
Glycobiology, 24, 2014
|
|
6EXP
 
 | | Crystal structure of the SIRV3 AcrID1 (gp02) anti-CRISPR protein | | 分子名称: | SIRV3 AcrID1 (gp02) anti-CRISPR protein | | 著者 | He, F, Bhoobalan-Chitty, Y, Van, L.B, Kjeldsen, A.L, Dedola, M, Makarova, K.S, Koonin, E.V, Brodersen, D.E, Peng, X. | | 登録日 | 2017-11-08 | | 公開日 | 2018-01-31 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.93 Å) | | 主引用文献 | Anti-CRISPR proteins encoded by archaeal lytic viruses inhibit subtype I-D immunity.
Nat Microbiol, 3, 2018
|
|
3WCS
 
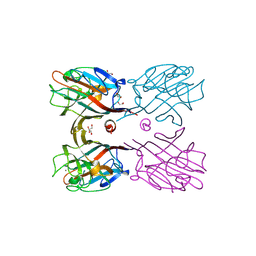 | | Crystal structure of plant lectin (ligand-bound form) | | 分子名称: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose, ... | | 著者 | Nagae, M, Yamaguchi, Y. | | 登録日 | 2013-05-31 | | 公開日 | 2014-04-09 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Phytohemagglutinin from Phaseolus vulgaris (PHA-E) displays a novel glycan recognition mode using a common legume lectin fold
Glycobiology, 24, 2014
|
|
7PZO
 
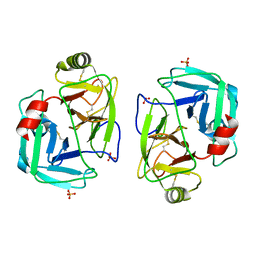 | | mite allergen Der p 3 from Dermatophagoides pteronyssinus | | 分子名称: | SULFATE ION, mite allergen Der p 3 | | 著者 | Timofeev, V.I, Shevtsov, M.B, Abramchik, Y.A, Mikheeva, O.O, Kostromina, M.A, Lykoshin, D.D, Zayats, E.A, Zavriev, S.K, Esipov, R.S, Kuranova, I.P. | | 登録日 | 2021-10-13 | | 公開日 | 2022-11-02 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | Structural plasticity and thermal stability of the histone-like protein from Spiroplasma melliferum are due to phenylalanine insertions into the conservative scaffold.
J.Biomol.Struct.Dyn., 36, 2018
|
|
8J3S
 
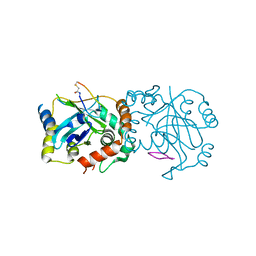 | | Complex structure of human cytomegalovirus protease and a macrocyclic peptide ligand | | 分子名称: | Assemblin, PHE-ILE-THR-GLY-HIS-TYR-TRP-VAL-ARG-PHE-LEU-PRO-CYS-GLY | | 著者 | Yoshida, S, Sako, Y, Nikaido, E, Ueda, T, Kozono, I, Ichihashi, Y, Nakahashi, A, Onishi, M, Yamatsu, Y, Kato, T, Nishikawa, J, Tachibana, Y. | | 登録日 | 2023-04-18 | | 公開日 | 2023-11-08 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (3.09 Å) | | 主引用文献 | Peptide-to-Small Molecule: Discovery of Non-Covalent, Active-Site Inhibitors of beta-Herpesvirus Proteases.
Acs Med.Chem.Lett., 14, 2023
|
|
8J3T
 
 | | Complex structure of human cytomegalovirus protease and a non-covalent small-molecule ligand | | 分子名称: | (4R)-1-[1-[(S)-[1-cyclopentyl-3-(2-methylphenyl)pyrazol-4-yl]-(4-methylphenyl)methyl]-2-oxidanylidene-pyridin-3-yl]-3-methyl-2-oxidanylidene-N-(3-oxidanylidene-2-azabicyclo[2.2.2]octan-4-yl)imidazolidine-4-carboxamide, Assemblin | | 著者 | Yoshida, S, Sako, Y, Nikaido, E, Ueda, T, Kozono, I, Ichihashi, Y, Nakahashi, A, Onishi, M, Yamatsu, Y, Kato, T, Nishikawa, J, Tachibana, Y. | | 登録日 | 2023-04-18 | | 公開日 | 2023-11-08 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Peptide-to-Small Molecule: Discovery of Non-Covalent, Active-Site Inhibitors of beta-Herpesvirus Proteases.
Acs Med.Chem.Lett., 14, 2023
|
|
2D07
 
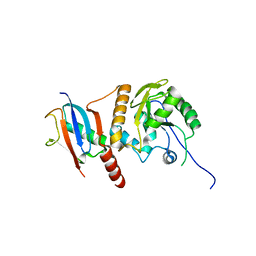 | | Crystal Structure of SUMO-3-modified Thymine-DNA Glycosylase | | 分子名称: | G/T mismatch-specific thymine DNA glycosylase, Ubiquitin-like protein SMT3B | | 著者 | Baba, D, Maita, N, Jee, J.G, Uchimura, Y, Saitoh, H, Sugasawa, K, Hanaoka, F, Tochio, H, Hiroaki, H, Shirakawa, M. | | 登録日 | 2005-07-26 | | 公開日 | 2006-06-06 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Crystal Structure of SUMO-3-modified Thymine-DNA Glycosylase
J.Mol.Biol., 359, 2006
|
|
3VYJ
 
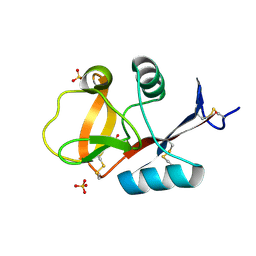 | | Crystal structure of C-type lectin domain of murine dendritic cell inhibitory receptor 2 (apo form) | | 分子名称: | C-type lectin domain family 4, member a4, SULFATE ION | | 著者 | Nagae, M, Yamanaka, K, Hanashima, S, Ikeda, A, Satoh, T, Matsumoto, N, Yamamoto, K, Yamaguchi, Y. | | 登録日 | 2012-09-26 | | 公開日 | 2013-10-02 | | 最終更新日 | 2013-12-11 | | 実験手法 | X-RAY DIFFRACTION (2.15 Å) | | 主引用文献 | Recognition of Bisecting N-Acetylglucosamine: STRUCTURAL BASIS FOR ASYMMETRIC INTERACTION WITH THE MOUSE LECTIN DENDRITIC CELL INHIBITORY RECEPTOR 2
J.Biol.Chem., 288, 2013
|
|
7YU1
 
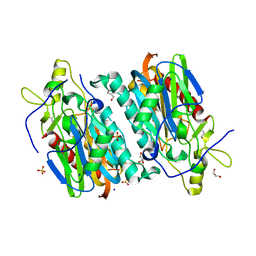 | |
7YU0
 
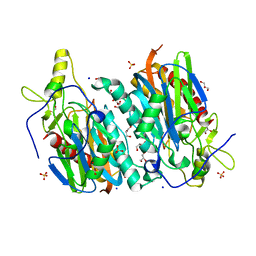 | |
6EEQ
 
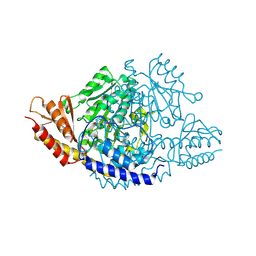 | | Crystal structure of Rhodiola rosea 4-hydroxyphenylacetaldehyde synthase | | 分子名称: | 4-hydroxyphenylacetaldehyde synthase | | 著者 | Torrens-Spence, M.P, Chiang, Y, Smith, T, Vicent, M.A, Wang, Y, Weng, J.K. | | 登録日 | 2018-08-15 | | 公開日 | 2018-09-19 | | 最終更新日 | 2020-06-03 | | 実験手法 | X-RAY DIFFRACTION (2.600086 Å) | | 主引用文献 | Structural basis for divergent and convergent evolution of catalytic machineries in plant aromatic amino acid decarboxylase proteins.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
8HUL
 
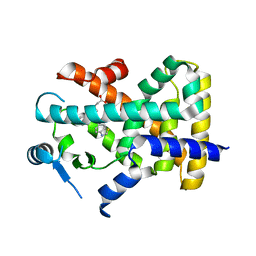 | | X-ray structure of human PPAR delta ligand binding domain-lanifibranor co-crystals obtained by co-crystallization | | 分子名称: | 4-[1-(1,3-benzothiazol-6-ylsulfonyl)-5-chloro-indol-2-yl]butanoic acid, Peroxisome proliferator-activated receptor delta | | 著者 | Kamata, S, Honda, A, Machida, Y, Uchii, K, Shiiyama, Y, Masuda, R, Oyama, T, Ishii, I. | | 登録日 | 2022-12-24 | | 公開日 | 2023-08-09 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.461 Å) | | 主引用文献 | Functional and Structural Insights into the Human PPAR alpha / delta / gamma Targeting Preferences of Anti-NASH Investigational Drugs, Lanifibranor, Seladelpar, and Elafibranor.
Antioxidants, 12, 2023
|
|
8HUN
 
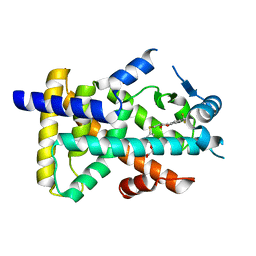 | | X-ray structure of human PPAR alpha ligand binding domain-seladelpar co-crystals obtained by cross-seeding | | 分子名称: | GLYCEROL, Peroxisome proliferator-activated receptor alpha, Seladelpar | | 著者 | Kamata, S, Honda, A, Machida, Y, Uchii, K, Shiiyama, Y, Masuda, R, Iino, S, Oyama, T, Ishii, I. | | 登録日 | 2022-12-24 | | 公開日 | 2023-08-09 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.01 Å) | | 主引用文献 | Functional and Structural Insights into the Human PPAR alpha / delta / gamma Targeting Preferences of Anti-NASH Investigational Drugs, Lanifibranor, Seladelpar, and Elafibranor.
Antioxidants, 12, 2023
|
|
8HUP
 
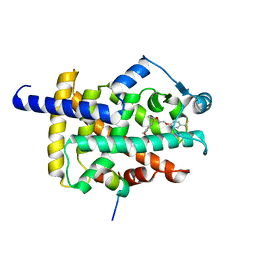 | | X-ray structure of human PPAR gamma ligand binding domain-seladelpar-SRC1 coactivator peptide co-crystals obtained by co-crystallization | | 分子名称: | 15-meric peptide from Nuclear receptor coactivator 1, Isoform 1 of Peroxisome proliferator-activated receptor gamma, Seladelpar | | 著者 | Kamata, S, Honda, A, Machida, Y, Uchii, K, Shiiyama, Y, Masuda, R, Oyama, T, Ishii, I. | | 登録日 | 2022-12-24 | | 公開日 | 2023-08-09 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.36 Å) | | 主引用文献 | Functional and Structural Insights into the Human PPAR alpha / delta / gamma Targeting Preferences of Anti-NASH Investigational Drugs, Lanifibranor, Seladelpar, and Elafibranor.
Antioxidants, 12, 2023
|
|
8HUM
 
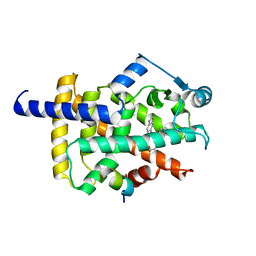 | | X-ray structure of human PPAR gamma ligand binding domain-lanifibranor-SRC1 coactivator peptide co-crystals obtained by co-crystallization | | 分子名称: | 15-meric peptide from Nuclear receptor coactivator 1, 4-[1-(1,3-benzothiazol-6-ylsulfonyl)-5-chloro-indol-2-yl]butanoic acid, Isoform 1 of Peroxisome proliferator-activated receptor gamma | | 著者 | Kamata, S, Honda, A, Machida, Y, Uchii, K, Shiiyama, Y, Masuda, R, Oyama, T, Ishii, I. | | 登録日 | 2022-12-24 | | 公開日 | 2023-08-09 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.29 Å) | | 主引用文献 | Functional and Structural Insights into the Human PPAR alpha / delta / gamma Targeting Preferences of Anti-NASH Investigational Drugs, Lanifibranor, Seladelpar, and Elafibranor.
Antioxidants, 12, 2023
|
|
