8HX9
 
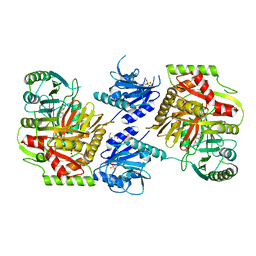 | | Crystal structure of 4-amino-4-deoxychorismate synthase from Streptomyces venezuelae with chorismate | | Descriptor: | (3R,4R)-3-[(1-carboxyethenyl)oxy]-4-hydroxycyclohexa-1,5-diene-1-carboxylic acid, 4-amino-4-deoxychorismate synthase, FORMIC ACID, ... | | Authors: | Nakamichi, Y, Watanabe, M. | | Deposit date: | 2023-01-04 | | Release date: | 2023-10-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Structural basis for the allosteric pathway of 4-amino-4-deoxychorismate synthase.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
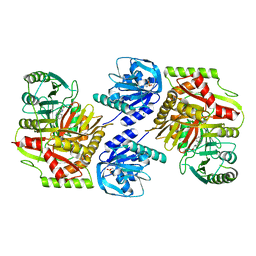
8HX8
 
 | |
6MY1
 
 | | Solution structure of gomesin at 278 K | | Descriptor: | gomesin | | Authors: | Chin, Y.K.-Y, Deplazes, E. | | Deposit date: | 2018-10-31 | | Release date: | 2019-11-06 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | The unusual conformation of cross-strand disulfide bonds is critical to the stability of beta-hairpin peptides.
Proteins, 88, 2020
|
|
6MY2
 
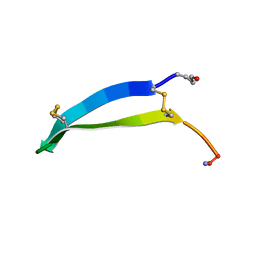 | | Solution structure of gomesin at 298 K | | Descriptor: | gomesin | | Authors: | Chin, Y.K.-Y, Deplazes, E. | | Deposit date: | 2018-10-31 | | Release date: | 2019-11-06 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | The unusual conformation of cross-strand disulfide bonds is critical to the stability of beta-hairpin peptides.
Proteins, 88, 2020
|
|
6MY3
 
 | | Solution structure of gomesin at 310K | | Descriptor: | gomesin | | Authors: | Chin, Y.K.-Y, Deplazes, E. | | Deposit date: | 2018-11-01 | | Release date: | 2019-11-06 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | The unusual conformation of cross-strand disulfide bonds is critical to the stability of beta-hairpin peptides.
Proteins, 88, 2020
|
|
7DHL
 
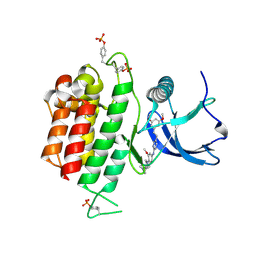 | | Crystal structure of FGFR3 in complex with pyrimidine derivative | | Descriptor: | 5-[2-(3,5-dimethoxyphenyl)ethyl]-N-[3-methoxy-4-[4-(4-methylpiperazin-1-yl)piperidin-1-yl]phenyl]pyrimidin-2-amine, Fibroblast growth factor receptor 3 | | Authors: | Echizen, Y, Tateishi, Y, Amano, Y. | | Deposit date: | 2020-11-16 | | Release date: | 2021-02-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Synthesis and structure-activity relationships of pyrimidine derivatives as potent and orally active FGFR3 inhibitors with both increased systemic exposure and enhanced in vitro potency.
Bioorg.Med.Chem., 33, 2021
|
|
8JOR
 
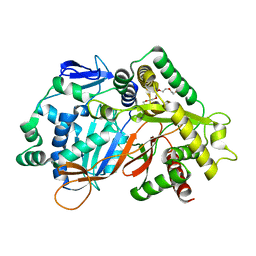 | | Structure of an acyltransferase involved in mannosylerythritol lipid formation from Pseudozyma tsukubaensis in type A crystal | | Descriptor: | Acyltransferase, PENTAETHYLENE GLYCOL | | Authors: | Nakamichi, Y, Saika, A, Watanabe, M, Fujii, T, Morita, T. | | Deposit date: | 2023-06-08 | | Release date: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structural identification of catalytic His158 of PtMAC2p from Pseudozyma tsukubaensis , an acyltransferase involved in mannosylerythritol lipids formation.
Front Bioeng Biotechnol, 11, 2023
|
|
8JOS
 
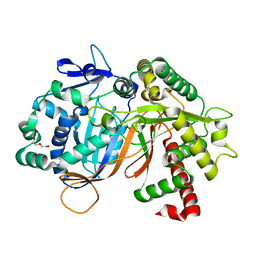 | | Structure of an acyltransferase involved in mannosylerythritol lipid formation from Pseudozyma tsukubaensis in type B crystal | | Descriptor: | Acyltransferase, CHLORIDE ION, TRIETHYLENE GLYCOL | | Authors: | Nakamichi, Y, Saika, A, Watanabe, M, Fujii, T, Morita, T. | | Deposit date: | 2023-06-08 | | Release date: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Structural identification of catalytic His158 of PtMAC2p from Pseudozyma tsukubaensis , an acyltransferase involved in mannosylerythritol lipids formation.
Front Bioeng Biotechnol, 11, 2023
|
|
7F2W
 
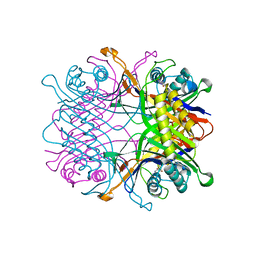 | | TbUox in complex with uric acid | | Descriptor: | URIC ACID, Uricase | | Authors: | Chiu, Y.C, Hsu, T.S, Huang, C.Y, Hsu, C.H. | | Deposit date: | 2021-06-14 | | Release date: | 2022-05-04 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Structural and biochemical insights into a hyperthermostable urate oxidase from Thermobispora bispora for hyperuricemia and gout therapy.
Int.J.Biol.Macromol., 188, 2021
|
|
5WLX
 
 | |
7DTM
 
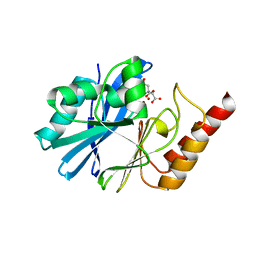 | |
7DTN
 
 | |
2N8F
 
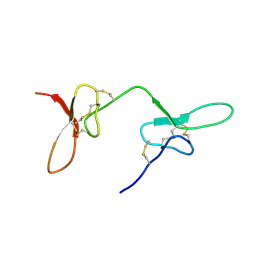 | |
3GYX
 
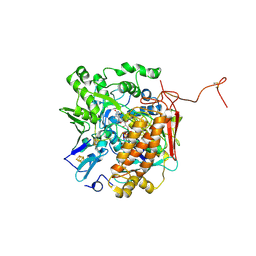 | | Crystal structure of adenylylsulfate reductase from Desulfovibrio gigas | | Descriptor: | Adenylylsulfate Reductase, FLAVIN-ADENINE DINUCLEOTIDE, IRON/SULFUR CLUSTER | | Authors: | Chiang, Y.-L, Hsieh, Y.-C, Liu, E.-H, Liu, M.-Y, Chen, C.-J. | | Deposit date: | 2009-04-06 | | Release date: | 2009-12-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structure of Adenylylsulfate reductase from Desulfovibrio gigas suggests a potential self-regulation mechanism involving the C terminus of the beta-subunit
J.Bacteriol., 191, 2009
|
|
2NBC
 
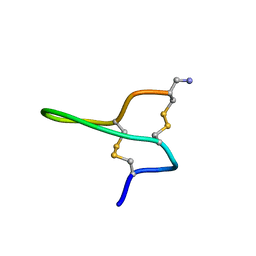 | |
2N8K
 
 | | Chemical Shift Assignments and Structure Determination for spider toxin, U33-theraphotoxin-Cg1c | | Descriptor: | U33-theraphotoxin-Cg1b | | Authors: | Chin, Y.K.-Y, Pineda, S.S, Mobli, M, King, G.F. | | Deposit date: | 2015-10-20 | | Release date: | 2016-08-31 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structural venomics reveals evolution of a complex venom by duplication and diversification of an ancient peptide-encoding gene.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6KRL
 
 | | Crystal structure of GH30 xylanase B from Talaromyces cellulolyticus expressed by Pichia pastoris | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, ... | | Authors: | Nakamichi, Y, Watanabe, M, Inoue, H. | | Deposit date: | 2019-08-22 | | Release date: | 2020-06-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.601 Å) | | Cite: | Substrate recognition by a bifunctional GH30-7 xylanase B from Talaromyces cellulolyticus.
Febs Open Bio, 10, 2020
|
|
6LFT
 
 | | Poa1p S30A mutant in complex with ADP-ribose | | Descriptor: | ACETATE ION, ADENOSINE-5-DIPHOSPHORIBOSE, ADP-ribose 1''-phosphate phosphatase | | Authors: | Chiu, Y.C, Hsu, C.H. | | Deposit date: | 2019-12-03 | | Release date: | 2020-12-09 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Expanding the Substrate Specificity of Macro Domains toward 3''-Isomer of O-Acetyl-ADP-ribose
Acs Catalysis, 11, 2021
|
|
6LFS
 
 | | Poa1p H23A mutant in complex with ADP-ribose | | Descriptor: | 3-CYCLOHEXYL-1-PROPYLSULFONIC ACID, ADENOSINE-5-DIPHOSPHORIBOSE, ADP-ribose 1''-phosphate phosphatase | | Authors: | Chiu, Y.C, Hsu, C.H. | | Deposit date: | 2019-12-03 | | Release date: | 2020-12-09 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Expanding the Substrate Specificity of Macro Domains toward 3''-Isomer of O-Acetyl-ADP-ribose
Acs Catalysis, 11, 2021
|
|
6LFQ
 
 | | Crystal structure of Poa1p in apo form | | Descriptor: | ADP-ribose 1''-phosphate phosphatase, GLYCEROL | | Authors: | Chiu, Y.C, Hsu, C.H. | | Deposit date: | 2019-12-03 | | Release date: | 2020-12-09 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.359 Å) | | Cite: | Expanding the Substrate Specificity of Macro Domains toward 3''-Isomer of O-Acetyl-ADP-ribose
Acs Catalysis, 11, 2021
|
|
5X6Y
 
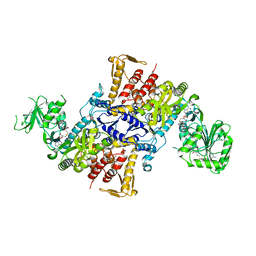 | |
6LFR
 
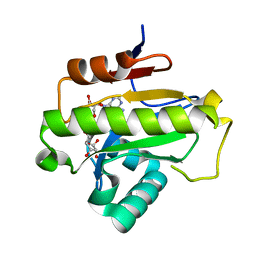 | | Poa1p in complex with ADP-ribose | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, ADP-ribose 1''-phosphate phosphatase | | Authors: | Chiu, Y.C, Hsu, C.H. | | Deposit date: | 2019-12-03 | | Release date: | 2020-12-09 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Expanding the Substrate Specificity of Macro Domains toward 3''-Isomer of O-Acetyl-ADP-ribose
Acs Catalysis, 11, 2021
|
|
2M32
 
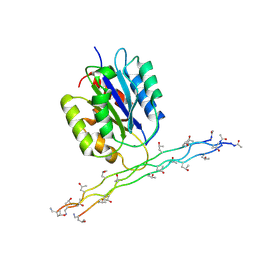 | | Alpha-1 integrin I-domain in complex with GLOGEN triple helical peptide | | Descriptor: | GLOGEN peptide, Integrin alpha-1, MAGNESIUM ION | | Authors: | Chin, Y, Headey, S, Mohanty, B, McEwan, P, Swarbrick, J, Mulhern, T, Emsley, J, Simpson, J, Scanlon, M. | | Deposit date: | 2013-01-07 | | Release date: | 2013-11-06 | | Last modified: | 2014-02-12 | | Method: | SOLUTION NMR | | Cite: | The Structure of Integrin alpha 1I Domain in Complex with a Collagen-mimetic Peptide.
J.Biol.Chem., 288, 2013
|
|
6M5Z
 
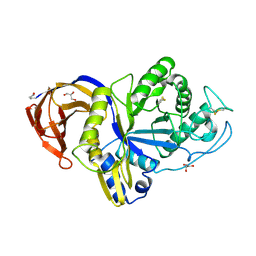 | | Catalytic domain of GH30 xylanase C from Talaromyces cellulolyticus | | Descriptor: | ACETATE ION, GH30 Xylanase C, GLYCEROL, ... | | Authors: | Nakamichi, Y, Watanabe, M, Inoue, H. | | Deposit date: | 2020-03-12 | | Release date: | 2021-01-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of GH30-7 endoxylanase C from the filamentous fungus Talaromyces cellulolyticus.
Acta Crystallogr.,Sect.F, 76, 2020
|
|
3VSN
 
 | | The crystal structure of novel chondroition lyase ODV-E66, baculovirus envelope protein | | Descriptor: | GLYCEROL, IODIDE ION, Occlusion-derived virus envelope protein E66 | | Authors: | Kawaguchi, Y, Sugiura, N, Kimata, K, Kimura, M, Kakuta, Y. | | Deposit date: | 2012-04-27 | | Release date: | 2013-05-22 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The crystal structure of novel chondroition lyase ODV-E66, baculovirus envelope protein
To be Published
|
|
