7EEN
 
 | |
3WXO
 
 | | Crystal structure of isoniazid bound KatG catalase peroxidase from Synechococcus elongatus PCC7942 | | 分子名称: | Catalase-peroxidase, PROTOPORPHYRIN IX CONTAINING FE, SODIUM ION, ... | | 著者 | Wada, K, Tada, T, Kamachi, S. | | 登録日 | 2014-08-04 | | 公開日 | 2015-01-21 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.12 Å) | | 主引用文献 | The crystal structure of isoniazid-bound KatG catalase-peroxidase from Synechococcus elongatus PCC7942.
Febs J., 282, 2015
|
|
3WNU
 
 | | The crystal structure of catalase-peroxidase, KatG, from Synechococcus PCC7942 | | 分子名称: | Catalase-peroxidase, HEME B/C, SODIUM ION | | 著者 | Tada, T, Wada, K, Kamachi, S. | | 登録日 | 2013-12-17 | | 公開日 | 2014-03-12 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | The 2.2 angstrom resolution structure of the catalase-peroxidase KatG from Synechococcus elongatus PCC7942.
Acta Crystallogr.,Sect.F, 70, 2014
|
|
2N3G
 
 | |
7BPF
 
 | | Structure of L-threoninol nucleic acid - RNA complex | | 分子名称: | L-aTNA (3'-(*GP*CP*AP*GP*CP*AP*GP*C)-1'), RNA (5'-R(*GP*CP*UP*GP*CP*(5BU)P*GP*C)-3') | | 著者 | Kamiya, Y, Satoh, T, Kodama, A, Suzuki, T, Uchiyama, S, Kato, K, Asanuma, H. | | 登録日 | 2020-03-22 | | 公開日 | 2020-11-25 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Intrastrand backbone-nucleobase interactions stabilize unwound right-handed helical structures of heteroduplexes of L-aTNA/RNA and SNA/RNA
Commun Chem, 2020
|
|
7UJZ
 
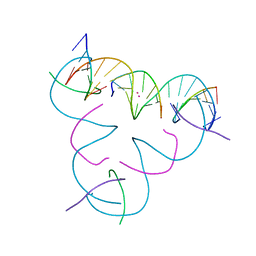 | | [T:Cd2+/Hg2+:T--pH 11] Metal-mediated DNA base pair in tensegrity triangle | | 分子名称: | CADMIUM ION, DNA (5'-D(*GP*AP*GP*CP*AP*GP*CP*CP*TP*GP*TP*TP*TP*GP*GP*AP*CP*AP*TP*CP*A)-3'), DNA (5'-D(P*CP*CP*AP*TP*AP*CP*A)-3'), ... | | 著者 | Lu, B, Vecchioni, S, Seeman, N.C, Sha, R, Ohayon, Y.P. | | 登録日 | 2022-03-31 | | 公開日 | 2023-04-05 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (3.94 Å) | | 主引用文献 | Heterobimetallic Base Pair Programming in Designer 3D DNA Crystals.
J.Am.Chem.Soc., 145, 2023
|
|
7UK0
 
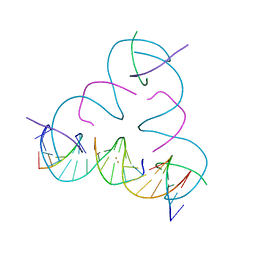 | | [T:Au+:T--pH 11 w/ HgCl2] Metal-mediated DNA base pair in tensegrity triangle | | 分子名称: | DNA (5'-D(*GP*AP*GP*CP*AP*GP*CP*CP*TP*GP*TP*TP*TP*GP*GP*AP*CP*AP*TP*CP*A)-3'), DNA (5'-D(P*CP*CP*AP*TP*AP*CP*A)-3'), DNA (5'-D(P*CP*TP*GP*AP*TP*GP*T)-3'), ... | | 著者 | Lu, B, Vecchioni, S, Seeman, N.C, Sha, R, Ohayon, Y.P. | | 登録日 | 2022-03-31 | | 公開日 | 2023-04-05 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (4.36 Å) | | 主引用文献 | Heterobimetallic Base Pair Programming in Designer 3D DNA Crystals.
J.Am.Chem.Soc., 145, 2023
|
|
7UL8
 
 | | [T:Au+:T--pH 11] Metal-mediated DNA base pair in tensegrity triangle | | 分子名称: | DNA (5'-D(*GP*AP*GP*CP*AP*GP*CP*CP*TP*GP*TP*TP*TP*GP*GP*AP*CP*AP*TP*CP*A)-3'), DNA (5'-D(P*CP*CP*AP*TP*AP*CP*A)-3'), DNA (5'-D(P*CP*TP*GP*AP*TP*GP*T)-3'), ... | | 著者 | Lu, B, Vecchioni, S, Seeman, N.C, Sha, R, Ohayon, Y.P. | | 登録日 | 2022-04-04 | | 公開日 | 2023-04-05 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (4.22 Å) | | 主引用文献 | Metal-Mediated DNA Nanotechnology in 3D: Structural Library by Templated Diffraction.
Adv Mater, 35, 2023
|
|
6GYM
 
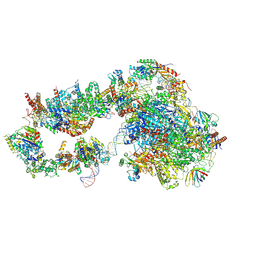 | | Structure of a yeast closed complex with distorted DNA (CCdist) | | 分子名称: | DNA-directed RNA polymerase II subunit RPB1, DNA-directed RNA polymerase II subunit RPB11, DNA-directed RNA polymerase II subunit RPB2, ... | | 著者 | Dienemann, C, Schwalb, B, Schilbach, S, Cramer, P. | | 登録日 | 2018-06-30 | | 公開日 | 2018-12-05 | | 最終更新日 | 2024-05-15 | | 実験手法 | ELECTRON MICROSCOPY (6.7 Å) | | 主引用文献 | Promoter Distortion and Opening in the RNA Polymerase II Cleft.
Mol. Cell, 73, 2019
|
|
1XKT
 
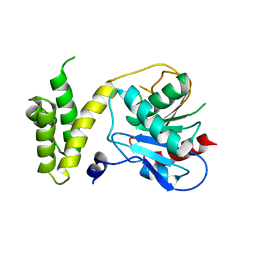 | | Human fatty acid synthase: Structure and substrate selectivity of the thioesterase domain | | 分子名称: | fatty acid synthase | | 著者 | Chakravarty, B, Gu, Z, Chirala, S.S, Wakil, S.J, Quiocho, F.A. | | 登録日 | 2004-09-29 | | 公開日 | 2004-10-26 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Human fatty acid synthase: structure and substrate selectivity of the thioesterase domain.
Proc.Natl.Acad.Sci.Usa, 101, 2004
|
|
3X16
 
 | |
5ZNO
 
 | | Crystal structure of PET-degrading cutinase Cut190 S176A/S226P/R228S/ mutant in Ca(2+)-bound state | | 分子名称: | Alpha/beta hydrolase family protein, CALCIUM ION, GLYCEROL | | 著者 | Numoto, N, Inaba, S, Yamagami, Y, Kamiya, N, Bekker, G.J, Ishii, K, Uchiyama, S, Kawai, F, Ito, N, Oda, M. | | 登録日 | 2018-04-10 | | 公開日 | 2018-09-12 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.60264349 Å) | | 主引用文献 | Structural Dynamics of the PET-Degrading Cutinase-like Enzyme from Saccharomonospora viridis AHK190 in Substrate-Bound States Elucidates the Ca2+-Driven Catalytic Cycle.
Biochemistry, 57, 2018
|
|
3ZFP
 
 | | Crystal structure of product-like, processed N-terminal protease Npro with internal His-Tag | | 分子名称: | CHLORIDE ION, MONOTHIOGLYCEROL, N-TERMINAL PROTEASE NPRO | | 著者 | Zogg, T, Sponring, M, Schindler, S, Koll, M, Schneider, R, Brandstetter, H, Auer, B. | | 登録日 | 2012-12-12 | | 公開日 | 2013-05-15 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.25 Å) | | 主引用文献 | Crystal Structures of the Viral Protease Npro Imply Distinct Roles for the Catalytic Water in Catalysis
Structure, 21, 2013
|
|
3ZFT
 
 | | Crystal structure of product-like, processed N-terminal protease Npro at pH 3 | | 分子名称: | CHLORIDE ION, MONOTHIOGLYCEROL, N-TERMINAL PROTEASE NPRO | | 著者 | Zogg, T, Sponring, M, Schindler, S, Koll, M, Schneider, R, Brandstetter, H, Auer, B. | | 登録日 | 2012-12-12 | | 公開日 | 2013-05-15 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Crystal Structures of the Viral Protease Npro Imply Distinct Roles for the Catalytic Water in Catalysis
Structure, 21, 2013
|
|
3VS9
 
 | | Crystal structure of type III PKS ArsC mutant | | 分子名称: | SODIUM ION, TETRAETHYLENE GLYCOL, Type III polyketide synthase | | 著者 | Satou, R, Miyanaga, A, Ozawa, H, Funa, N, Miyazono, K, Tanokura, M, Ohnishi, Y, Horinouchi, S. | | 登録日 | 2012-04-23 | | 公開日 | 2013-04-24 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.99 Å) | | 主引用文献 | Structural basis for cyclization specificity of two Azotobacter type III polyketide synthases: a single amino acid substitution reverses their cyclization specificity
J.Biol.Chem., 288, 2013
|
|
5ZRS
 
 | | Crystal structure of PET-degrading cutinase Cut190 S176A/S226P/R228S mutant in monoethyl adipate bound state | | 分子名称: | 6-ethoxy-6-oxohexanoic acid, Alpha/beta hydrolase family protein, CALCIUM ION, ... | | 著者 | Numoto, N, Kamiya, N, Bekker, G.J, Yamagami, Y, Inaba, S, Ishii, K, Uchiyama, S, Kawai, F, Ito, N, Oda, M. | | 登録日 | 2018-04-25 | | 公開日 | 2018-09-12 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Structural Dynamics of the PET-Degrading Cutinase-like Enzyme from Saccharomonospora viridis AHK190 in Substrate-Bound States Elucidates the Ca2+-Driven Catalytic Cycle.
Biochemistry, 57, 2018
|
|
3ZFU
 
 | | Crystal structure of substrate-like, unprocessed N-terminal protease Npro mutant S169P with sulphate | | 分子名称: | MONOTHIOGLYCEROL, N-TERMINAL PROTEASE NPRO, SULFATE ION | | 著者 | Zogg, T, Sponring, M, Schindler, S, Koll, M, Schneider, R, Brandstetter, H, Auer, B. | | 登録日 | 2012-12-12 | | 公開日 | 2013-05-15 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.76 Å) | | 主引用文献 | Crystal Structures of the Viral Protease Npro Imply Distinct Roles for the Catalytic Water in Catalysis
Structure, 21, 2013
|
|
3ZFO
 
 | | Crystal structure of substrate-like, unprocessed N-terminal protease Npro mutant S169P | | 分子名称: | CHLORIDE ION, HYDROXIDE ION, MONOTHIOGLYCEROL, ... | | 著者 | Zogg, T, Sponring, M, Schindler, S, Koll, M, Schneider, R, Brandstetter, H, Auer, B. | | 登録日 | 2012-12-12 | | 公開日 | 2013-05-15 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.01 Å) | | 主引用文献 | Crystal Structures of the Viral Protease Npro Imply Distinct Roles for the Catalytic Water in Catalysis
Structure, 21, 2013
|
|
3RGW
 
 | | Crystal structure at 1.5 A resolution of an H2-reduced, O2-tolerant hydrogenase from Ralstonia eutropha unmasks a novel iron-sulfur cluster | | 分子名称: | FE3-S4 CLUSTER, FE4-S3 CLUSTER, IRON/SULFUR CLUSTER, ... | | 著者 | Scheerer, P, Fritsch, J, Frielingsdorf, S, Kroschinsky, S, Friedrich, B, Lenz, O, Spahn, C.M.T. | | 登録日 | 2011-04-10 | | 公開日 | 2011-10-26 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | The crystal structure of an oxygen-tolerant hydrogenase uncovers a novel iron-sulphur centre.
Nature, 479, 2011
|
|
2N3H
 
 | |
5ZQT
 
 | | Crystal structure of Oryza sativa hexokinase 6 | | 分子名称: | Hexokinase-6, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | 著者 | Matsudaira, K, Mochizuki, S, Yoshida, H, Kamitori, S, Akimitsu, K. | | 登録日 | 2018-04-20 | | 公開日 | 2019-04-24 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.84 Å) | | 主引用文献 | Crystal structure of Oryza sativa hexokinase 6
To Be Published
|
|
2N3F
 
 | |
3ZFR
 
 | | Crystal structure of product-like, processed N-terminal protease Npro with iridium | | 分子名称: | HYDROXIDE ION, IRIDIUM (III) ION, MONOTHIOGLYCEROL, ... | | 著者 | Zogg, T, Sponring, M, Schindler, S, Koll, M, Schneider, R, Brandstetter, H, Auer, B. | | 登録日 | 2012-12-12 | | 公開日 | 2013-05-15 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Crystal Structures of the Viral Protease Npro Imply Distinct Roles for the Catalytic Water in Catalysis
Structure, 21, 2013
|
|
3ZFQ
 
 | | Crystal structure of product-like, processed N-terminal protease Npro with mercury | | 分子名称: | MERCURY (II) ION, MONOTHIOGLYCEROL, N-TERMINAL PROTEASE NPRO | | 著者 | Zogg, T, Sponring, M, Schindler, S, Koll, M, Schneider, R, Brandstetter, H, Auer, B. | | 登録日 | 2012-12-12 | | 公開日 | 2013-05-15 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.65 Å) | | 主引用文献 | Crystal Structures of the Viral Protease Npro Imply Distinct Roles for the Catalytic Water in Catalysis
Structure, 21, 2013
|
|
3ZFN
 
 | | Crystal structure of product-like, processed N-terminal protease Npro | | 分子名称: | CHLORIDE ION, MONOTHIOGLYCEROL, N-TERMINAL PROTEASE NPRO | | 著者 | Zogg, T, Sponring, M, Schindler, S, Koll, M, Schneider, R, Brandstetter, H, Auer, B. | | 登録日 | 2012-12-12 | | 公開日 | 2013-05-15 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Crystal Structures of the Viral Protease Npro Imply Distinct Roles for the Catalytic Water in Catalysis
Structure, 21, 2013
|
|
