2FL0
 
 | | Oxidized (All ferric) form of the Azotobacter vinelandii bacterioferritin | | Descriptor: | Bacterioferritin, FE (II) ION, MAGNESIUM ION, ... | | Authors: | Swartz, L, Kunchinskas, K, Li, H, Poulos, T.L, Lanzilotta, W.N. | | Deposit date: | 2006-01-05 | | Release date: | 2006-04-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Redox-Dependent Structural Changes in the Azotobacter vinelandii Bacterioferritin: New Insights into the Ferroxidase and Iron Transport Mechanism(,).
Biochemistry, 45, 2006
|
|
3VYG
 
 | | Crystal structure of Thiocyanate hydrolase mutant R136W | | Descriptor: | COBALT (III) ION, L(+)-TARTARIC ACID, Thiocyanate hydrolase subunit alpha, ... | | Authors: | Yamanaka, Y, Sato, M, Arakawa, T, Namima, S, Hori, S, Ohtaki, A, Noguchi, K, Katayama, Y, Yohda, M, Odaka, M. | | Deposit date: | 2012-09-25 | | Release date: | 2013-11-13 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Effects of argnine residue around the substrate pocket on the substrate specificity of thiocyanate hydrolase
To be published
|
|
1HZG
 
 | | CRYSTAL STRUCTURE OF THE INACTIVE C866S MUTANT OF THE CATALYTIC DOMAIN OF E. COLI CYTOTOXIC NECROTIZING FACTOR 1 | | Descriptor: | CYTOTOXIC NECROTIZING FACTOR 1, PHOSPHATE ION | | Authors: | Buetow, L, Flatau, G, Chiu, K, Boquet, P, Ghosh, P. | | Deposit date: | 2001-01-24 | | Release date: | 2001-07-04 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Structure of the Rho-activating domain of Escherichia coli cytotoxic necrotizing factor 1.
Nat.Struct.Biol., 8, 2001
|
|
1HQ0
 
 | | CRYSTAL STRUCTURE OF THE CATALYTIC DOMAIN OF E.COLI CYTOTOXIC NECROTIZING FACTOR TYPE 1 | | Descriptor: | CYTOTOXIC NECROTIZING FACTOR 1, PHOSPHATE ION | | Authors: | Buetow, L, Flatau, G, Chiu, K, Boquet, P, Ghosh, P. | | Deposit date: | 2000-12-13 | | Release date: | 2001-07-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Structure of the Rho-activating domain of Escherichia coli cytotoxic necrotizing factor 1.
Nat.Struct.Biol., 8, 2001
|
|
2ZYK
 
 | | Crystal structure of cyclo/maltodextrin-binding protein complexed with gamma-cyclodextrin | | Descriptor: | Cyclooctakis-(1-4)-(alpha-D-glucopyranose), Solute-binding protein | | Authors: | Tonozuka, T, Sogawa, A, Yamada, M, Matsumoto, N, Yoshida, H, Kamitori, S, Ichikawa, K, Mizuno, M, Nishikawa, A, Sakano, Y. | | Deposit date: | 2009-01-26 | | Release date: | 2009-02-10 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for cyclodextrin recognition by Thermoactinomyces vulgaris cyclo/maltodextrin-binding protein
Febs J., 274, 2007
|
|
3RDN
 
 | | NMR STRUCTURE OF THE N-TERMINAL DOMAIN WITH A LINKER PORTION OF ANTARCTIC EEL POUT ANTIFREEZE PROTEIN RD3, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | ANTIFREEZE PROTEIN RD3 TYPE III | | Authors: | Miura, K, Ohgiya, S, Hoshino, T, Nemoto, N, Hikichi, K, Tsuda, S. | | Deposit date: | 1998-02-24 | | Release date: | 1999-02-23 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural basis for the binding of a globular antifreeze protein to ice.
Nature, 384, 1996
|
|
3B0D
 
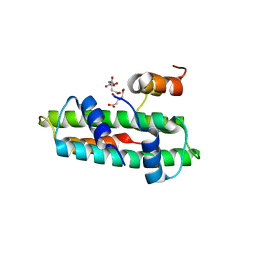 | | Crystal structure of the chicken CENP-T histone fold/CENP-W complex, crystal form II | | Descriptor: | CITRIC ACID, Centromere protein T, Centromere protein W | | Authors: | Nishino, T, Takeuchi, K, Gascoigne, K.E, Suzuki, A, Hori, T, Oyama, T, Morikawa, K, Cheeseman, I.M, Fukagawa, T. | | Deposit date: | 2011-06-08 | | Release date: | 2012-03-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.197 Å) | | Cite: | CENP-T-W-S-X Forms a Unique Centromeric Chromatin Structure with a Histone-like Fold.
Cell(Cambridge,Mass.), 148, 2012
|
|
1JL8
 
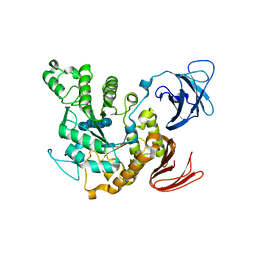 | | Complex of alpha-amylase II (TVA II) from Thermoactinomyces vulgaris R-47 with beta-cyclodextrin based on a co-crystallization with methyl beta-cyclodextrin | | Descriptor: | ALPHA-AMYLASE II, Cycloheptakis-(1-4)-(alpha-D-glucopyranose) | | Authors: | Yokota, T, Tonozuka, T, Shimura, Y, Ichikawa, K, Kamitori, S, Sakano, Y. | | Deposit date: | 2001-07-16 | | Release date: | 2001-08-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structures of Thermoactinomyces vulgaris R-47 alpha-amylase II complexed with substrate analogues.
Biosci.Biotechnol.Biochem., 65, 2001
|
|
3A8L
 
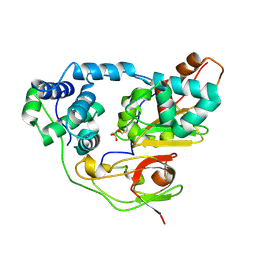 | | Crystal structure of photo-activation state of Nitrile Hydratase mutant S113A | | Descriptor: | FE (III) ION, Nitrile hydratase subunit alpha, Nitrile hydratase subunit beta | | Authors: | Yamanaka, Y, Hashimoto, K, Ohtaki, A, Noguchi, K, Yohda, M, Odaka, M. | | Deposit date: | 2009-10-06 | | Release date: | 2010-04-14 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Kinetic and structural studies on roles of the serine ligand and a strictly conserved tyrosine residue in nitrile hydratase
J.Biol.Inorg.Chem., 15, 2010
|
|
3A1I
 
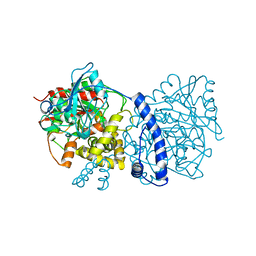 | | Crystal structure of Rhodococcus sp. N-771 Amidase complexed with Benzamide | | Descriptor: | Amidase, BENZAMIDE | | Authors: | Ohtaki, A, Noguchi, K, Sato, Y, Murata, K, Odaka, M, Yohda, M. | | Deposit date: | 2009-04-03 | | Release date: | 2009-10-27 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Structure and Characterization of Amidase from Rhodococcus sp. N-771: Insight into the Molecular Mechanism of Substrate Recognition
Biochim.Biophys.Acta, 2009
|
|
3A8O
 
 | | Crystal structure of Nitrile Hydratase complexed with Trimethylacetamide | | Descriptor: | 2,2-dimethylpropanamide, FE (III) ION, Nitrile hydratase subunit alpha, ... | | Authors: | Yamanaka, Y, Hashimoto, K, Ohtaki, A, Noguchi, K, Yohda, M, Odaka, M. | | Deposit date: | 2009-10-07 | | Release date: | 2010-04-14 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Kinetic and structural studies on roles of the serine ligand and a strictly conserved tyrosine residue in nitrile hydratase
J.Biol.Inorg.Chem., 15, 2010
|
|
3A8H
 
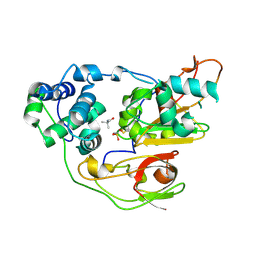 | | Crystal structure of Nitrile Hydratase mutant S113A complexed with Trimethylacetamide | | Descriptor: | 2,2-dimethylpropanamide, FE (III) ION, Nitrile hydratase subunit alpha, ... | | Authors: | Yamanaka, Y, Hashimoto, K, Ohtaki, A, Noguchi, K, Yohda, M, Odaka, M. | | Deposit date: | 2009-10-06 | | Release date: | 2010-04-14 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Kinetic and structural studies on roles of the serine ligand and a strictly conserved tyrosine residue in nitrile hydratase
J.Biol.Inorg.Chem., 15, 2010
|
|
1QWF
 
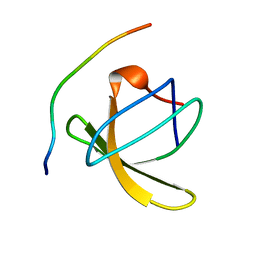 | | C-SRC SH3 DOMAIN COMPLEXED WITH LIGAND VSL12 | | Descriptor: | TYROSINE-PROTEIN KINASE TRANSFORMING PROTEIN SRC, VAL-SER-LEU-ALA-ARG-ARG-PRO-LEU-PRO-PRO-LEU-PRO | | Authors: | Feng, S, Chiyoshi, K, Rickles, R.J, Schreiber, S.L. | | Deposit date: | 1995-11-09 | | Release date: | 1996-03-08 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Specific interactions outside the proline-rich core of two classes of Src homology 3 ligands.
Proc.Natl.Acad.Sci.USA, 92, 1995
|
|
1QWE
 
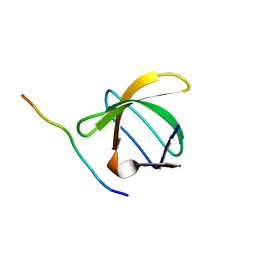 | | C-SRC SH3 DOMAIN COMPLEXED WITH LIGAND APP12 | | Descriptor: | ALA-PRO-PRO-LEU-PRO-PRO-ARG-ASN-ARG-PRO-ARG-LEU, TYROSINE-PROTEIN KINASE TRANSFORMING PROTEIN SRC | | Authors: | Feng, S, Chiyoshi, K, Rickles, R.J, Schreiber, S.L. | | Deposit date: | 1995-11-09 | | Release date: | 1996-03-08 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Specific interactions outside the proline-rich core of two classes of Src homology 3 ligands.
Proc.Natl.Acad.Sci.USA, 92, 1995
|
|
3VPL
 
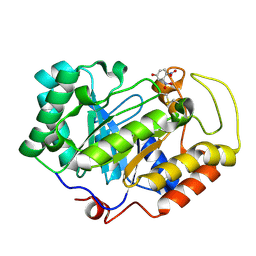 | | Crystal structure of a 2-fluoroxylotriosyl complex of the Vibrio sp. AX-4 Beta-1,3-xylanase | | Descriptor: | 3,4-dinitrophenol, Beta-1,3-xylanase XYL4, beta-D-xylopyranose-(1-3)-beta-D-xylopyranose-(1-3)-2-deoxy-2-fluoro-beta-D-xylopyranose, ... | | Authors: | Watanabe, N, Sakaguchi, K. | | Deposit date: | 2012-03-05 | | Release date: | 2013-03-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | The crystal structure of a 2-fluoroxylotriosyl complex of the Vibrio sp. AX-4 beta-1,3-xylanase at 1.2 A resolution
To be Published
|
|
7MF3
 
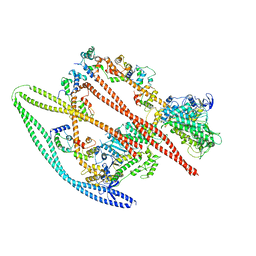 | | Structure of the autoinhibited state of smooth muscle myosin-2 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Myosin light polypeptide 6, ... | | Authors: | Heissler, S.M, Arora, A.S, Billington, N, Sellers, J.R, Chinthalapudi, K. | | Deposit date: | 2021-04-08 | | Release date: | 2022-01-05 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-EM structure of the autoinhibited state of myosin-2.
Sci Adv, 7, 2021
|
|
2ONC
 
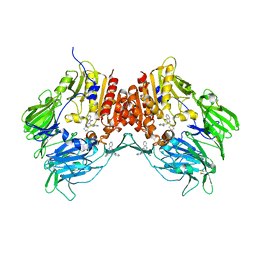 | | Crystal structure of human DPP-4 | | Descriptor: | 2-({2-[(3R)-3-AMINOPIPERIDIN-1-YL]-4-OXOQUINAZOLIN-3(4H)-YL}METHYL)BENZONITRILE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Feng, J, Zhang, Z, Wallace, M.B, Stafford, J.A, Kaldor, S.W, Kassel, D.B, Navre, M, Shi, L, Skene, R.J, Asakawa, T, Takeuchi, K, Xu, R, Webb, D.R, Gwaltney, S.L. | | Deposit date: | 2007-01-23 | | Release date: | 2008-03-04 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Discovery of alogliptin: a potent, selective, bioavailable, and efficacious inhibitor of dipeptidyl peptidase IV.
J.Med.Chem., 50, 2007
|
|
1SMY
 
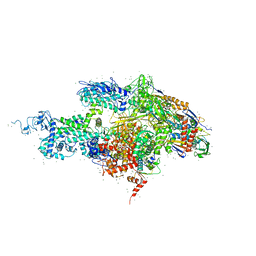 | | Structural basis for transcription regulation by alarmone ppGpp | | Descriptor: | DNA-directed RNA polymerase alpha chain, DNA-directed RNA polymerase beta chain, DNA-directed RNA polymerase beta' chain, ... | | Authors: | Artsimovitch, I, Patlan, V, Sekine, S, Vassylyeva, M.N, Hosaka, T, Ochi, K, Yokoyama, S, Vassylyev, D.G, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-03-10 | | Release date: | 2004-05-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis for transcription regulation by alarmone ppGpp
Cell(Cambridge,Mass.), 117, 2004
|
|
1GBS
 
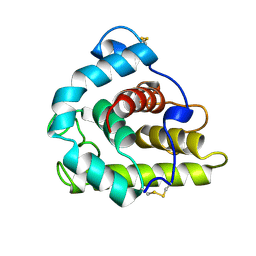 | |
2PDD
 
 | | THE HIGH RESOLUTION STRUCTURE OF THE PERIPHERAL SUBUNIT-BINDING DOMAIN OF DIHYDROLIPOAMIDE ACETYLTRANSFERASE FROM THE PYRUVATE DEHYDROGENASE MULTIENZYME COMPLEX OF BACILLUS STEAROTHERMOPHILUS | | Descriptor: | DIHYDROLIPOAMIDE ACETYLTRANSFERASE | | Authors: | Kalia, Y.N, Brocklehurst, S.M, Hipps, D.S, Appella, E, Sakaguchi, K, Perham, R.N. | | Deposit date: | 1992-11-25 | | Release date: | 1994-12-20 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | The high-resolution structure of the peripheral subunit-binding domain of dihydrolipoamide acetyltransferase from the pyruvate dehydrogenase multienzyme complex of Bacillus stearothermophilus.
J.Mol.Biol., 230, 1993
|
|
3CI7
 
 | | Crystal structure of a simplified BPTI containing 20 alanines | | Descriptor: | SULFATE ION, bovine pancreatic trypsin inhibitor | | Authors: | Islam, M.M, Sohya, S, Noguchi, K, Yohda, M, Kuroda, Y. | | Deposit date: | 2008-03-11 | | Release date: | 2008-10-21 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structure of an extensively simplified variant of bovine pancreatic trypsin inhibitor in which over one-third of the residues are alanines
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
2PDE
 
 | | THE HIGH RESOLUTION STRUCTURE OF THE PERIPHERAL SUBUNIT-BINDING DOMAIN OF DIHYDROLIPOAMIDE ACETYLTRANSFERASE FROM THE PYRUVATE DEHYDROGENASE MULTIENZYME COMPLEX OF BACILLUS STEAROTHERMOPHILUS | | Descriptor: | DIHYDROLIPOAMIDE ACETYLTRANSFERASE | | Authors: | Kalia, Y.N, Brocklehurst, S.M, Hipps, D.S, Appella, E, Sakaguchi, K, Perham, R.N. | | Deposit date: | 1992-11-25 | | Release date: | 1994-12-20 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | The high-resolution structure of the peripheral subunit-binding domain of dihydrolipoamide acetyltransferase from the pyruvate dehydrogenase multienzyme complex of Bacillus stearothermophilus.
J.Mol.Biol., 230, 1993
|
|
2XO8
 
 | | Crystal Structure of Myosin-2 in Complex with Tribromodichloropseudilin | | Descriptor: | 2,4-DICHLORO-6-(3,4,5-TRIBROMO-1H-PYRROL-2-YL)PHENOL, ADP METAVANADATE, MAGNESIUM ION, ... | | Authors: | Preller, M, Chinthalapudi, K, Martin, R, Knoelker, H.J, Manstein, D.J. | | Deposit date: | 2010-08-10 | | Release date: | 2011-05-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Inhibition of Myosin ATPase Activity by Halogenated Pseudilins: A Structure-Activity Study.
J.Med.Chem., 54, 2011
|
|
1WZK
 
 | | Thermoactinomyces vulgaris R-47 alpha-amylase II (TVA II) mutatnt D465N | | Descriptor: | Alpha-amylase II, CALCIUM ION | | Authors: | Mizuno, M, Ichikawa, K, Tonozuka, T, Ohtaki, A, Shimura, Y, Kamitori, S, Nishikawa, A, Sakano, Y. | | Deposit date: | 2005-03-06 | | Release date: | 2005-03-22 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Mutagenesis and Structural Analysis of Thermoactinomyces vulgaris R-47 alpha-Amylase II (TVA II)
To be Published
|
|
1WZL
 
 | | Thermoactinomyces vulgaris R-47 alpha-amylase II (TVA II) mutatnt R469L | | Descriptor: | Alpha-amylase II, CALCIUM ION | | Authors: | Mizuno, M, Ichikawa, K, Tonozuka, T, Ohtaki, A, Shimura, Y, Kamitori, S, Nishikawa, A, Sakano, Y. | | Deposit date: | 2005-03-06 | | Release date: | 2005-03-22 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Mutagenesis and Structural Analysis of Thermoactinomyces vulgaris R-47 alpha-Amylase II (TVA II)
To be Published
|
|
