1CVO
 
 | |
1CXZ
 
 | | CRYSTAL STRUCTURE OF HUMAN RHOA COMPLEXED WITH THE EFFECTOR DOMAIN OF THE PROTEIN KINASE PKN/PRK1 | | Descriptor: | 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE, MAGNESIUM ION, PROTEIN (HIS-TAGGED TRANSFORMING PROTEIN RHOA(0-181)), ... | | Authors: | Maesaki, R, Ihara, K, Shimizu, T, Kuroda, S, Kaibuchi, K, Hakoshima, T. | | Deposit date: | 1999-08-31 | | Release date: | 1999-10-08 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The structural basis of Rho effector recognition revealed by the crystal structure of human RhoA complexed with the effector domain of PKN/PRK1.
Mol.Cell, 4, 1999
|
|
1DPF
 
 | | CRYSTAL STRUCTURE OF A MG-FREE FORM OF RHOA COMPLEXED WITH GDP | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, RHOA | | Authors: | Shimizu, T, Ihara, K, Maesaki, R, Kuroda, S, Kaibuchi, K, Hakoshima, T. | | Deposit date: | 1999-12-27 | | Release date: | 2000-06-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | An open conformation of switch I revealed by the crystal structure of a Mg2+-free form of RHOA complexed with GDP. Implications for the GDP/GTP exchange mechanism.
J.Biol.Chem., 275, 2000
|
|
1CL4
 
 | |
6JSF
 
 | | Crystal Structure of BACE1 in complex with N-(3-((4S,5S)-2-amino-4-methyl-5-phenyl-5,6-dihydro-4H-1,3-thiazin-4-yl)-4-fluorophenyl)-5-(fluoromethoxy)pyrazine-2-carboxamide | | Descriptor: | Beta-secretase 1, DIMETHYL SULFOXIDE, IODIDE ION, ... | | Authors: | Fujimoto, K, Matsuoka, E, Asada, N, Tadano, G, Yamamoto, T, Nakahara, K, Fuchino, K, Ito, H, Kanegawa, N, Moechars, D, Gijsen, H.J.M, Kusakabe, K.I. | | Deposit date: | 2019-04-08 | | Release date: | 2019-08-28 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure-Based Design of Selective beta-Site Amyloid Precursor Protein Cleaving Enzyme 1 (BACE1) Inhibitors: Targeting the Flap to Gain Selectivity over BACE2.
J.Med.Chem., 62, 2019
|
|
8IVR
 
 | | Methyl and Fluorine Effects in Novel Orally Bioavailable Keap1/Nrf2 PPI Inhibitor for Treatment of Chronic Kidney Disease | | Descriptor: | (2R,3S)-2-methyl-3-(4-methylphenyl)-3-[2-(5,6,7,8-tetrahydronaphthalen-2-yl)ethanoylamino]propanoic acid, Kelch-like ECH-associated protein 1, SULFATE ION | | Authors: | Nomura, A, Yamaguchi, K, Adachi, T. | | Deposit date: | 2023-03-28 | | Release date: | 2023-06-14 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Methyl and Fluorine Effects in Novel Orally Bioavailable Keap1-Nrf2 PPI Inhibitor.
Acs Med.Chem.Lett., 14, 2023
|
|
8IXS
 
 | | Methyl and Fluorine Effects in Novel Orally Bioavailable Keap1/Nrf2 PPI Inhibitor for Treatment of Chronic Kidney Disease | | Descriptor: | (2R,3S)-3-[[(2S)-2-fluoranyl-2-(5,6,7,8-tetrahydronaphthalen-2-yl)ethanoyl]amino]-2-methyl-3-(4-methylphenyl)propanoic acid, Kelch-like ECH-associated protein 1, SULFATE ION | | Authors: | Nomura, A, Yamaguchi, K, Adachi, T. | | Deposit date: | 2023-04-03 | | Release date: | 2023-06-14 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Methyl and Fluorine Effects in Novel Orally Bioavailable Keap1-Nrf2 PPI Inhibitor.
Acs Med.Chem.Lett., 14, 2023
|
|
8IVG
 
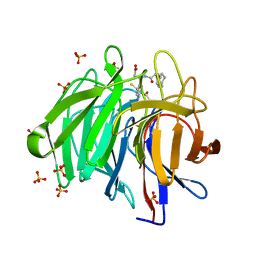 | | Methyl and Fluorine Effects in Novel Orally Bioavailable Keap1/Nrf2 PPI Inhibitor for Treatment of Chronic Kidney Disease | | Descriptor: | (3S)-3-(4-methylphenyl)-3-[2-(5,6,7,8-tetrahydronaphthalen-2-yl)ethanoylamino]propanoic acid, 2,3-DIHYDROXY-1,4-DITHIOBUTANE, Kelch-like ECH-associated protein 1, ... | | Authors: | Otake, K, Ubukata, M, Nagahashi, N, Ogawa, N, Hantani, Y, Hantani, R, Adachi, T, Nomura, A, Yamaguchi, K, Maekawa, M, Mamada, H, Motomura, T, Sato, M, Harada, K. | | Deposit date: | 2023-03-27 | | Release date: | 2023-06-14 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Methyl and Fluorine Effects in Novel Orally Bioavailable Keap1-Nrf2 PPI Inhibitor.
Acs Med.Chem.Lett., 14, 2023
|
|
6A22
 
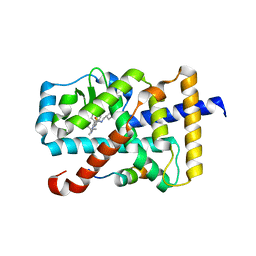 | | Ternary complex of Human ROR gamma Ligand Binding Domain With Compound T. | | Descriptor: | 2-[2-[1-~{tert}-butyl-5-(4-methoxyphenyl)pyrazol-4-yl]-1,3-thiazol-4-yl]-~{N}-(oxan-4-ylmethyl)ethanamide, Nuclear receptor ROR-gamma, Nuclear receptor corepressor 2 | | Authors: | Noguchi, M, Nomura, A, Doi, S, Yamaguchi, K, Adachi, T. | | Deposit date: | 2018-06-08 | | Release date: | 2018-12-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Ternary crystal structure of human ROR gamma ligand-binding-domain, an inhibitor and corepressor peptide provides a new insight into corepressor interaction
Sci Rep, 8, 2018
|
|
4WKR
 
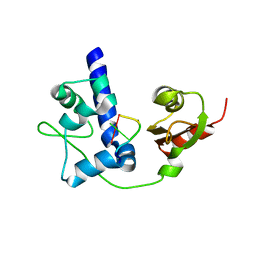 | | LaRP7 wrapping up the 3' hairpin of 7SK non-coding RNA (302-332) | | Descriptor: | 7SK GGHP4 (300-332), La-related protein 7 | | Authors: | Uchikawa, E, Natchiar, K.S, Han, X, Proux, F, Roblin, P, Zhang, E, Durand, A, Klaholz, B.P, Dock-Bregeon, A.-C. | | Deposit date: | 2014-10-03 | | Release date: | 2015-03-18 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural insight into the mechanism of stabilization of the 7SK small nuclear RNA by LARP7.
Nucleic Acids Res., 43, 2015
|
|
4WJO
 
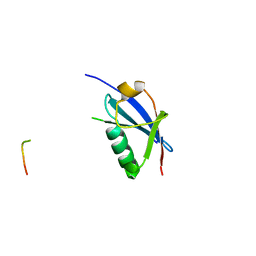 | | Crystal Structure of SUMO1 in complex with PML | | Descriptor: | Protein PML, Small ubiquitin-related modifier 1 | | Authors: | Cappadocia, L, Mascle, X.H, Bourdeau, V, Tremblay-Belzile, S, Chaker-Margot, M, Lussier-Price, M, Wada, J, Sakaguchi, K, Aubry, M, Ferbeyre, G, Omichinski, J.G. | | Deposit date: | 2014-10-01 | | Release date: | 2014-12-31 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Structural and Functional Characterization of the Phosphorylation-Dependent Interaction between PML and SUMO1.
Structure, 23, 2015
|
|
4WJN
 
 | | Crystal structure of SUMO1 in complex with phosphorylated PML | | Descriptor: | Protein PML, Small ubiquitin-related modifier 1 | | Authors: | Cappadocia, L, Mascle, X.H, Bourdeau, V, Tremblay-Belzile, S, Chaker-Margot, M, Lussier-Price, M, Wada, J, Sakaguchi, K, Aubry, M, Ferbeyre, G, Omichinski, J.G. | | Deposit date: | 2014-10-01 | | Release date: | 2014-12-31 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural and Functional Characterization of the Phosphorylation-Dependent Interaction between PML and SUMO1.
Structure, 23, 2015
|
|
4WJP
 
 | | Crystal Structure of SUMO1 in complex with phosphorylated Daxx | | Descriptor: | Daxx, Small ubiquitin-related modifier 1 | | Authors: | Cappadocia, L, Mascle, X.H, Bourdeau, V, Tremblay-Belzile, S, Chaker-Margot, M, Lussier-Price, M, Wada, J, Sakaguchi, K, Aubry, M, Ferbeyre, G, Omichinski, J.G. | | Deposit date: | 2014-10-01 | | Release date: | 2014-12-31 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural and Functional Characterization of the Phosphorylation-Dependent Interaction between PML and SUMO1.
Structure, 23, 2015
|
|
6JSG
 
 | | Crystal Structure of BACE1 in complex with N-{3-[(4S)-2-amino-4-methyl-5,6-dihydro-4H-1,3-thiazin-4-yl]-4-fluorophenyl}-5-chloropyridine-2-carboxamide | | Descriptor: | Beta-secretase 1, GLYCEROL, IODIDE ION, ... | | Authors: | Fujimoto, K, Matsuoka, E, Asada, N, Tadano, G, Yamamoto, T, Nakahara, K, Fuchino, K, Ito, H, Kanegawa, N, Moechars, D, Gijsen, H.J.M, Kusakabe, K.I. | | Deposit date: | 2019-04-08 | | Release date: | 2019-08-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure-Based Design of Selective beta-Site Amyloid Precursor Protein Cleaving Enzyme 1 (BACE1) Inhibitors: Targeting the Flap to Gain Selectivity over BACE2.
J.Med.Chem., 62, 2019
|
|
6JSZ
 
 | | BACE2 xaperone complex with N-{3-[(5R)-3-amino-5-methyl-9,9-dioxo-2,9lambda6-dithia-4-azaspiro[5.5]undec-3-en-5-yl]-4-fluorophenyl}-5-(fluoromethoxy)pyrazine-2-carboxamide | | Descriptor: | Beta-secretase 2, CHLORIDE ION, N-[3-[(5R)-3-azanyl-5-methyl-9,9-bis(oxidanylidene)-2,9$l^{6}-dithia-4-azaspiro[5.5]undec-3-en-5-yl]-4-fluoranyl-phenyl]-5-(fluoranylmethoxy)pyrazine-2-carboxamide, ... | | Authors: | Fujimoto, K, Matsuoka, E, Asada, N, Tadano, G, Yamamoto, T, Nakahara, K, Fuchino, K, Ito, H, Kanegawa, N, Moechars, D, Gijsen, H.J.M, Kusakabe, K.I. | | Deposit date: | 2019-04-08 | | Release date: | 2019-08-28 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Structure-Based Design of Selective beta-Site Amyloid Precursor Protein Cleaving Enzyme 1 (BACE1) Inhibitors: Targeting the Flap to Gain Selectivity over BACE2.
J.Med.Chem., 62, 2019
|
|
6JSN
 
 | | Crystal Structure of BACE1 in complex with N-{3-[(5R)-3-amino-5-methyl-9,9-dioxo-2,9lambda6-dithia-4-azaspiro[5.5]undec-3-en-5-yl]-4-fluorophenyl}-5-(fluoromethoxy)pyrazine-2-carboxamide | | Descriptor: | Beta-secretase 1, GLYCEROL, IODIDE ION, ... | | Authors: | Fujimoto, K, Matsuoka, E, Asada, N, Tadano, G, Yamamoto, T, Nakahara, K, Fuchino, K, Ito, H, Kanegawa, N, Moechars, D, Gijsen, H.J.M, Kusakabe, K.I. | | Deposit date: | 2019-04-08 | | Release date: | 2019-08-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure-Based Design of Selective beta-Site Amyloid Precursor Protein Cleaving Enzyme 1 (BACE1) Inhibitors: Targeting the Flap to Gain Selectivity over BACE2.
J.Med.Chem., 62, 2019
|
|
8WY1
 
 | | The structure of cyclization domain in cyclic beta-1,2-glucan synthase from Thermoanaerobacter italicus | | Descriptor: | Glycosyltransferase 36 | | Authors: | Tanaka, N, Saito, R, Kobayashi, K, Nakai, H, Kamo, S, Kuramochi, K, Taguchi, H, Nakajima, M, Masaike, T. | | Deposit date: | 2023-10-30 | | Release date: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.9 Å) | | Cite: | Functional and structural analysis of a cyclization domain in a cyclic beta-1,2-glucan synthase.
Appl.Microbiol.Biotechnol., 108, 2024
|
|
7X87
 
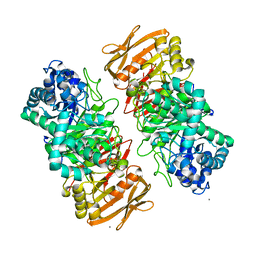 | | The complex structure of beta-1,2-glucosyltransferase from Ignavibacterium album with sophotetraose observed as sophorose | | Descriptor: | Beta-galactosidase, CALCIUM ION, beta-D-glucopyranose-(1-2)-beta-D-glucopyranose | | Authors: | Kobayashi, K, Shimizu, H, Tanaka, N, Kuramochi, K, Nakai, H, Nakajima, M, Taguchi, H. | | Deposit date: | 2022-03-11 | | Release date: | 2022-04-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Characterization and structural analyses of a novel glycosyltransferase acting on the beta-1,2-glucosidic linkages.
J Biol Chem, 298, 2022
|
|
7ZUW
 
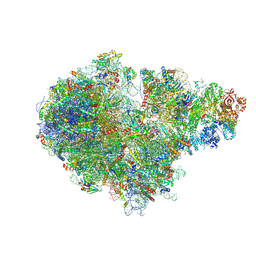 | | Structure of RQT (C1) bound to the stalled ribosome in a disome unit from S. cerevisiae | | Descriptor: | 18S ribosomal RNA, 25S ribosomal RNA, 40S ribosomal protein S0-A, ... | | Authors: | Best, K.M, Ikeuchi, K, Kater, L, Best, D.M, Musial, J, Matsuo, Y, Berninghausen, O, Becker, T, Inada, T, Beckmann, R. | | Deposit date: | 2022-05-13 | | Release date: | 2023-02-22 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structural basis for clearing of ribosome collisions by the RQT complex.
Nat Commun, 14, 2023
|
|
7ZPQ
 
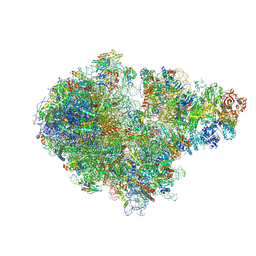 | | Structure of the RQT-bound 80S ribosome from S. cerevisiae (C1) | | Descriptor: | 18S ribosomal RNA, 25S ribosomal RNA, 40S ribosomal protein S0-A, ... | | Authors: | Best, K.M, Ikeuchi, K, Kater, L, Best, D.M, Musial, J, Matsuo, Y, Berninghausen, O, Becker, T, Inada, T, Beckmann, R. | | Deposit date: | 2022-04-28 | | Release date: | 2023-02-22 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.47 Å) | | Cite: | Structural basis for clearing of ribosome collisions by the RQT complex.
Nat Commun, 14, 2023
|
|
7ZS5
 
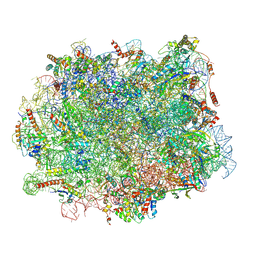 | | Structure of 60S ribosomal subunit from S. cerevisiae with eIF6 and tRNA | | Descriptor: | 25S ribosomal RNA, 5.8S ribosomal RNA, 5S ribosomal RNA, ... | | Authors: | Best, K.M, Ikeuchi, K, Kater, L, Best, D.M, Musial, J, Matsuo, Y, Berninghausen, O, Becker, T, Inada, T, Beckmann, R. | | Deposit date: | 2022-05-06 | | Release date: | 2023-02-22 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural basis for clearing of ribosome collisions by the RQT complex.
Nat Commun, 14, 2023
|
|
7ZRS
 
 | | Structure of the RQT-bound 80S ribosome from S. cerevisiae (C2) - composite map | | Descriptor: | 18S ribosomal RNA, 25S ribosomal RNA, 40S ribosomal protein S0-A, ... | | Authors: | Best, K.M, Ikeuchi, K, Kater, L, Best, D.M, Musial, J, Matsuo, Y, Berninghausen, O, Becker, T, Inada, T, Beckmann, R. | | Deposit date: | 2022-05-05 | | Release date: | 2023-02-22 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Structural basis for clearing of ribosome collisions by the RQT complex.
Nat Commun, 14, 2023
|
|
7ZUX
 
 | | Collided ribosome in a disome unit from S. cerevisiae | | Descriptor: | 18S ribosomal RNA, 25S ribosomal RNA, 40S ribosomal protein S0-A, ... | | Authors: | Best, K.M, Ikeuchi, K, Kater, L, Best, D.M, Musial, J, Matsuo, Y, Berninghausen, O, Becker, T, Inada, T, Beckmann, R. | | Deposit date: | 2022-05-13 | | Release date: | 2023-02-22 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Structural basis for clearing of ribosome collisions by the RQT complex.
Nat Commun, 14, 2023
|
|
5X8Q
 
 | | Crystal Structure of the mutant Human ROR gamma Ligand Binding Domain With rockogenin. | | Descriptor: | (1R,2S,4S,5'R,6R,7S,8R,9S,10R,12S,13S,16S,18S)-5',7,9,13-tetramethylspiro[5-oxapentacyclo[10.8.0.0^{2,9}.0^{4,8}.0^{13,18}]icosane-6,2'-oxane]-10,16-diol, Nuclear receptor ROR-gamma, Nuclear receptor corepressor 2 | | Authors: | Noguchi, M, Nomura, A, Murase, K, Doi, S, Yamaguchi, K, Adachi, T. | | Deposit date: | 2017-03-03 | | Release date: | 2017-06-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Ternary complex of human ROR gamma ligand-binding domain, inverse agonist and SMRT peptide shows a unique mechanism of corepressor recruitment
Genes Cells, 22, 2017
|
|
6KXX
 
 | | Human PPAR alpha ligand binding domain in complex with a synthetic agonist (compound A) | | Descriptor: | 1-(4-chlorophenyl)-6-methyl-3-propan-2-yl-pyrazolo[3,4-b]pyridine-4-carboxylic acid, PGC1alpha, Peroxisome proliferator-activated receptor alpha | | Authors: | Yoshida, T, Tachibana, K, Oki, H, Doi, M, Fukuda, S, Yuzuriha, T, Tabata, R, Ishimoto, K, Kawahara, K, Ohkubo, T, Miyachi, H, Doi, T. | | Deposit date: | 2019-09-14 | | Release date: | 2020-05-20 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural Basis for PPAR alpha Activation by 1H-pyrazolo-[3,4-b]pyridine Derivatives.
Sci Rep, 10, 2020
|
|
