1V7H
 
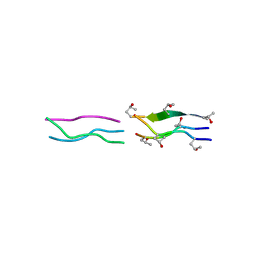 | | Crystal Structures of Collagen Model Peptides with Pro-Hyp-Gly Sequence at 1.26 A | | Descriptor: | Collagen like peptide | | Authors: | Okuyama, K, Hongo, C, Fukushima, R, Wu, G, Narita, H, Noguchi, K, Tanaka, Y, Nishino, N. | | Deposit date: | 2003-12-17 | | Release date: | 2004-08-03 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Crystal structures of collagen model peptides with Pro-Hyp-Gly repeating sequence at 1.26 A resolution: implications for proline ring puckering
Biopolymers, 76, 2004
|
|
1JJU
 
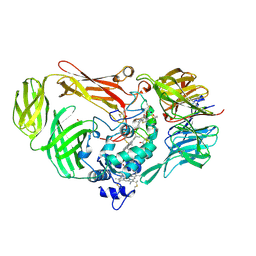 | | Structure of a Quinohemoprotein Amine Dehydrogenase with a Unique Redox Cofactor and Highly Unusual Crosslinking | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, QUINOHEMOPROTEIN AMINE DEHYDROGENASE, SODIUM ION, ... | | Authors: | Datta, S, Mori, Y, Takagi, K, Kawaguchi, K, Chen, Z.-W, Kano, K, Ikeda, T, Okajima, T, Kuroda, S, Tanizawa, K, Mathews, F.S. | | Deposit date: | 2001-07-09 | | Release date: | 2001-12-12 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structure of a quinohemoprotein amine dehydrogenase with an uncommon redox cofactor and highly unusual crosslinking.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
2CUO
 
 | | Collagen model peptide (PRO-PRO-GLY)9 | | Descriptor: | COLLAGEN MODEL PEPTIDE (PRO-PRO-GLY)9 | | Authors: | Hongo, C, Noguchi, K, Okuyama, K, Tanaka, Y, Nishino, N. | | Deposit date: | 2005-05-27 | | Release date: | 2005-06-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | Repetitive interactions observed in the crystal structure of a collagen-model peptide, [(Pro-Pro-Gly)9]3
J.Biochem.(Tokyo), 138, 2005
|
|
7D5U
 
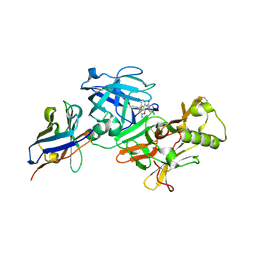 | | BACE2 xaperone complex with N-{3-[(9S)-7-amino-2,2-difluoro-9-(prop-1-yn-1-yl)-6-oxa-8-azaspiro[3.5]non-7-en-9-yl]-4-fluorophenyl}-5-cyanopyridine-2-carboxamide | | Descriptor: | Beta-secretase 2, N-[3-[(9S)-7-azanyl-2,2-bis(fluoranyl)-9-prop-1-ynyl-6-oxa-8-azaspiro[3.5]non-7-en-9-yl]-4-fluoranyl-phenyl]-5-cyano-pyridine-2-carboxamide, xaperone | | Authors: | Fujimoto, K, Yoshida, S, Tadano, G, Asada, N, Fuchino, K, Suzuki, S, Matsuoka, E, Yamamoto, T, Yamamoto, S, Ando, S, Kanegawa, N, Tonomura, Y, Ito, H, Moechars, D, Rombouts, F.J.R, Gijsen, H.J.M, Kusakabe, K.I. | | Deposit date: | 2020-09-28 | | Release date: | 2021-03-31 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Structure-Based Approaches to Improving Selectivity through Utilizing Explicit Water Molecules: Discovery of Selective beta-Secretase (BACE1) Inhibitors over BACE2.
J.Med.Chem., 64, 2021
|
|
7D2V
 
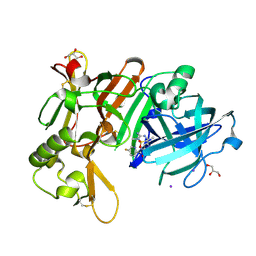 | | Crystal Structure of BACE1 in complex with N-{3-[(5R)-3-amino-2,5-dimethyl-1,1-dioxo-5,6-dihydro-2H-1lambda6,2,4-thiadiazin-5-yl]-4-fluorophenyl}-5-fluoropyridine-2-carboxamide | | Descriptor: | Beta-secretase 1, GLYCEROL, IODIDE ION, ... | | Authors: | Fujimoto, K, Yoshida, S, Tadano, G, Asada, N, Fuchino, K, Suzuki, S, Matsuoka, E, Yamamoto, T, Yamamoto, S, Ando, S, Kanegawa, N, Tonomura, Y, Ito, H, Moechars, D, Rombouts, F.J.R, Gijsen, H.J.M, Kusakabe, K.I. | | Deposit date: | 2020-09-17 | | Release date: | 2021-03-31 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-Based Approaches to Improving Selectivity through Utilizing Explicit Water Molecules: Discovery of Selective beta-Secretase (BACE1) Inhibitors over BACE2.
J.Med.Chem., 64, 2021
|
|
7D5B
 
 | | BACE2 xaperone complex with N-{3-[(5R)-3-amino-2,5-dimethyl-1,1-dioxo-5,6-dihydro-2H-1lambda6,2,4-thiadiazin-5-yl]-4-fluorophenyl}-5-fluoropyridine-2-carboxamide | | Descriptor: | 1,2-ETHANEDIOL, Beta-secretase 2, CHLORIDE ION, ... | | Authors: | Fujimoto, K, Yoshida, S, Tadano, G, Asada, N, Fuchino, K, Suzuki, S, Matsuoka, E, Yamamoto, T, Yamamoto, S, Ando, S, Kanegawa, N, Tonomura, Y, Ito, H, Moechars, D, Rombouts, F.J.R, Gijsen, H.J.M, Kusakabe, K.I. | | Deposit date: | 2020-09-25 | | Release date: | 2021-03-31 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | Structure-Based Approaches to Improving Selectivity through Utilizing Explicit Water Molecules: Discovery of Selective beta-Secretase (BACE1) Inhibitors over BACE2.
J.Med.Chem., 64, 2021
|
|
7D5A
 
 | | Crystal Structure of BACE1 in complex with N-{3-[(9S)-7-amino-2,2-difluoro-9-(prop-1-yn-1-yl)-6-oxa-8-azaspiro[3.5]non-7-en-9-yl]-4-fluorophenyl}-5-cyanopyridine-2-carboxamide | | Descriptor: | Beta-secretase 1, GLYCEROL, IODIDE ION, ... | | Authors: | Fujimoto, K, Yoshida, S, Tadano, G, Asada, N, Fuchino, K, Suzuki, S, Matsuoka, E, Yamamoto, T, Yamamoto, S, Ando, S, Kanegawa, N, Tonomura, Y, Ito, H, Moechars, D, Rombouts, F.J.R, Gijsen, H.J.M, Kusakabe, K.I. | | Deposit date: | 2020-09-25 | | Release date: | 2021-03-31 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure-Based Approaches to Improving Selectivity through Utilizing Explicit Water Molecules: Discovery of Selective beta-Secretase (BACE1) Inhibitors over BACE2.
J.Med.Chem., 64, 2021
|
|
7D2X
 
 | | Crystal Structure of BACE1 in complex with N-{3-[(4R)-2-amino-4-(prop-1-yn-1-yl)-5,6-dihydro-4H-1,3-oxazin-4-yl]-4-fluorophenyl}-5-cyanopyridine-2-carboxamide | | Descriptor: | Beta-secretase 1, GLYCEROL, IODIDE ION, ... | | Authors: | Fujimoto, K, Yoshida, S, Tadano, G, Asada, N, Fuchino, K, Suzuki, S, Matsuoka, E, Yamamoto, T, Yamamoto, S, Ando, S, Kanegawa, N, Tonomura, Y, Ito, H, Moechars, D, Rombouts, F.J.R, Gijsen, H.J.M, Kusakabe, K.I. | | Deposit date: | 2020-09-17 | | Release date: | 2021-03-31 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structure-Based Approaches to Improving Selectivity through Utilizing Explicit Water Molecules: Discovery of Selective beta-Secretase (BACE1) Inhibitors over BACE2.
J.Med.Chem., 64, 2021
|
|
2D42
 
 | | Crystal structure analysis of a non-toxic crystal protein from Bacillus thuringiensis | | Descriptor: | non-toxic crystal protein | | Authors: | Akiba, T, Higuchi, K, Mizuki, E, Ekino, K, Shin, T, Ohba, M, Kanai, R, Harata, K. | | Deposit date: | 2005-10-05 | | Release date: | 2006-01-17 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Nontoxic crystal protein from Bacillus thuringiensis demonstrates a remarkable structural similarity to beta-pore-forming toxins
Proteins, 63, 2006
|
|
7VPY
 
 | | Crystal structure of the neutralizing nanobody P86 against SARS-CoV-2 | | Descriptor: | 1,2-ETHANEDIOL, Nanobody, SULFATE ION | | Authors: | Maeda, R, Fujita, J, Konishi, Y, Kazuma, Y, Yamazaki, H, Anzai, I, Yamaguchi, K, Kasai, K, Nagata, K, Yamaoka, Y, Miyakawa, K, Ryo, A, Shirakawa, K, Makino, F, Matsuura, Y, Inoue, T, Imura, A, Namba, K, Takaori-Kondo, A. | | Deposit date: | 2021-10-18 | | Release date: | 2022-07-20 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A panel of nanobodies recognizing conserved hidden clefts of all SARS-CoV-2 spike variants including Omicron.
Commun Biol, 5, 2022
|
|
7VQ0
 
 | | Cryo-EM structure of the SARS-CoV-2 spike protein (2-up RBD) bound to neutralizing nanobodies P86 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Maeda, R, Fujita, J, Konishi, Y, Kazuma, Y, Yamazaki, H, Anzai, I, Yamaguchi, K, Kasai, K, Nagata, K, Yamaoka, Y, Miyakawa, K, Ryo, A, Shirakawa, K, Makino, F, Matsuura, Y, Inoue, T, Imura, A, Namba, K, Takaori-Kondo, A. | | Deposit date: | 2021-10-18 | | Release date: | 2022-07-20 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.03 Å) | | Cite: | A panel of nanobodies recognizing conserved hidden clefts of all SARS-CoV-2 spike variants including Omicron.
Commun Biol, 5, 2022
|
|
4BS1
 
 | | MuB is an AAAplus ATPase that forms helical filaments to control target selection for DNA transposition | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, TRANSCRIPTIONAL REGULATOR (NTRC FAMILY) | | Authors: | Mizuno, N, Dramicanin, M, Mizuuchi, M, Adam, J, Wang, Y, Han, Y.W, Yang, W, Steven, A.C, Mizuuchi, K, Ramon-Maiques, S. | | Deposit date: | 2013-06-06 | | Release date: | 2013-07-03 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (18 Å) | | Cite: | Mub is an Aaa+ ATPase that Forms Helical Filaments to Control Target Selection for DNA Transposition.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
5XT6
 
 | | A sulfur-transferring catalytic intermediate of SufS-SufU complex from Bacillus subtilis | | Descriptor: | 2-[(3-HYDROXY-2-METHYL-5-PHOSPHONOOXYMETHYL-PYRIDIN-4-YLMETHYL)-AMINO]-PROPIONIC ACID, Cysteine desulfurase SufS, ZINC ION, ... | | Authors: | Fujishiro, T, Kunichika, K, Takahashi, Y. | | Deposit date: | 2017-06-17 | | Release date: | 2017-12-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Zinc-Ligand Swapping Mediated Complex Formation and Sulfur Transfer between SufS and SufU for Iron-Sulfur Cluster Biogenesis in Bacillus subtilis
J. Am. Chem. Soc., 139, 2017
|
|
1WS8
 
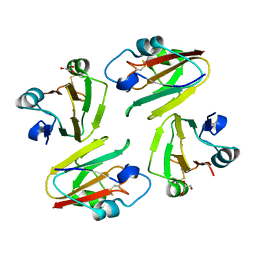 | | Crystal Structure of Mavicyanin from Cucurbita pepo medullosa (Zucchini) | | Descriptor: | COPPER (II) ION, GLYCEROL, mavicyanin | | Authors: | Xie, Y, Inoue, T, Miyamoto, Y, Matsumura, H, Kunishige, K, Yamaguchi, K, Nojini, M, Suzuki, S, Kai, Y. | | Deposit date: | 2004-11-02 | | Release date: | 2004-11-23 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural reorganization of the copper binding site involving Thr15 of mavicyanin from Cucurbita pepo medullosa (zucchini) upon reduction.
J.Biochem.(Tokyo), 137, 2005
|
|
1WS7
 
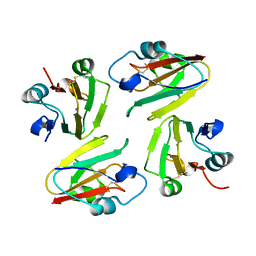 | | Crystal Structure of Mavicyanin from Cucurbita pepo medullosa (Zucchini) | | Descriptor: | COPPER (I) ION, Mavicyanin | | Authors: | Xie, Y, Inoue, T, Miyamoto, Y, Matsumura, H, Kataoka, K, Yamaguchi, K, Nojini, M, Suzuki, S, Kai, Y. | | Deposit date: | 2004-11-02 | | Release date: | 2004-11-23 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural reorganization of the copper binding site involving Thr15 of mavicyanin from Cucurbita pepo medullosa (zucchini) upon reduction.
J.Biochem.(Tokyo), 137, 2005
|
|
1IWC
 
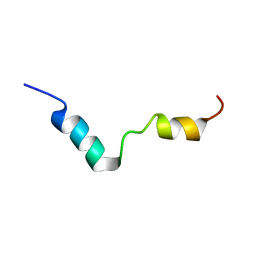 | | TFE-induded structure of the N-terminal domain of pig gastric H/K-ATPase | | Descriptor: | gastric H/K-ATPase | | Authors: | Fujitani, N, Kanagawa, M, Aizawa, T, Ohkubo, T, Kaya, S, Demura, M, Kawano, K, Taniguchi, K, Nitta, K. | | Deposit date: | 2002-05-02 | | Release date: | 2002-11-27 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Structure determination and conformational change induced by tyrosine phosphorylation of the N-terminal domain of the alpha-chain of pig gastric H+/K+-ATPase
Biochem.Biophys.Res.Commun., 300, 2003
|
|
1EQ5
 
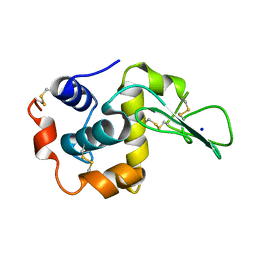 | | CRYSTAL STRUCTURES OF SALT BRIDGE MUTANTS OF HUMAN LYSOZYME | | Descriptor: | LYSOZYME, SODIUM ION | | Authors: | Takano, K, Tsuchimori, K, Yamagata, Y, Yutani, K. | | Deposit date: | 2000-04-03 | | Release date: | 2000-04-19 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Contribution of salt bridges near the surface of a protein to the conformational stability.
Biochemistry, 39, 2000
|
|
1EQE
 
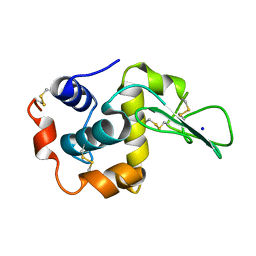 | | CRYSTAL STRUCTURES OF SALT BRIDGE MUTANTS OF HUMAN LYSOZYME | | Descriptor: | LYSOZYME, SODIUM ION | | Authors: | Takano, K, Tsuchimori, K, Yamagata, Y, Yutani, K. | | Deposit date: | 2000-04-04 | | Release date: | 2000-04-19 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Contribution of salt bridges near the surface of a protein to the conformational stability.
Biochemistry, 39, 2000
|
|
2D05
 
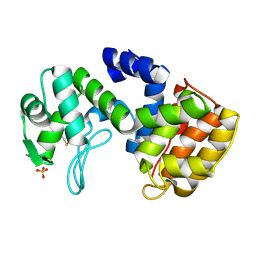 | | Chitosanase From Bacillus circulans mutant K218P | | Descriptor: | Chitosanase, SULFATE ION | | Authors: | Fukamizo, T, Amano, S, Yamaguchi, K, Yoshikawa, T, Katsumi, T, Saito, J, Suzuki, M, Miki, K, Nagata, Y, Ando, A. | | Deposit date: | 2005-07-25 | | Release date: | 2005-12-06 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Bacillus circulans MH-K1 Chitosanase: Amino Acid Residues Responsible for Substrate Binding
J.Biochem.(Tokyo), 138, 2005
|
|
1SMY
 
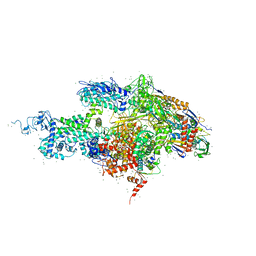 | | Structural basis for transcription regulation by alarmone ppGpp | | Descriptor: | DNA-directed RNA polymerase alpha chain, DNA-directed RNA polymerase beta chain, DNA-directed RNA polymerase beta' chain, ... | | Authors: | Artsimovitch, I, Patlan, V, Sekine, S, Vassylyeva, M.N, Hosaka, T, Ochi, K, Yokoyama, S, Vassylyev, D.G, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-03-10 | | Release date: | 2004-05-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis for transcription regulation by alarmone ppGpp
Cell(Cambridge,Mass.), 117, 2004
|
|
4Z9F
 
 | | Halohydrin hydrogen-halide-lyase, HheA | | Descriptor: | CHLORIDE ION, Halohydrin epoxidase A | | Authors: | Watanabe, F, Yu, F, Ohtaki, A, Yamanaka, Y, Noguchi, K, Yohda, M, Odaka, M. | | Deposit date: | 2015-04-10 | | Release date: | 2015-11-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structures of halohydrin hydrogen-halide-lyases from Corynebacterium sp. N-1074
Proteins, 83, 2015
|
|
7SSF
 
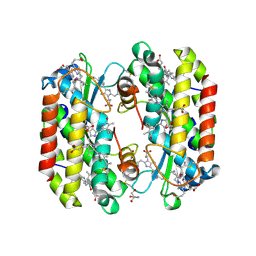 | | Light harvesting phycobiliprotein HaPE560 from the cryptophyte Hemiselmis andersenii CCMP644 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, DiCys-(15,16)-Dihydrobiliverdin, ... | | Authors: | Rathbone, H.W, Michie, K.A, Laos, A.L, Curmi, P.M.G. | | Deposit date: | 2021-11-10 | | Release date: | 2023-10-25 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Molecular dissection of the soluble photosynthetic antenna from the cryptophyte alga Hemiselmis andersenii
Commun Biol, 2023
|
|
7SUT
 
 | | Light harvesting phycobiliprotein HaPE645 from the cryptophyte Hemiselmis andersenii CCMP644 | | Descriptor: | (15,16)-DIHYDROBILIVERDIN (SINGLY LINKED), 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, ... | | Authors: | Rathbone, H.W, Michie, K.A, Laos, A.L, Curmi, P.M.G. | | Deposit date: | 2021-11-18 | | Release date: | 2023-10-25 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Molecular dissection of the soluble photosynthetic antenna from the cryptophyte alga Hemiselmis andersenii.
Commun Biol, 6, 2023
|
|
4ZU3
 
 | | Halohydrin hydrogen-halide-lyases, HheB | | Descriptor: | 3-hydroxypentanedinitrile, Halohydrin epoxidase B, MAGNESIUM ION, ... | | Authors: | Watanabe, F, Yu, F, Ohtaki, A, Yamanaka, Y, Noguchi, K, Yohda, M, Odaka, M. | | Deposit date: | 2015-05-15 | | Release date: | 2015-11-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of halohydrin hydrogen-halide-lyases from Corynebacterium sp. N-1074
Proteins, 83, 2015
|
|
2DUK
 
 | | Crystal structure of MS0616 | | Descriptor: | MS0616 | | Authors: | Hosaka, T, Nishino, A, Uchikubo, K.-T, Kishishita, S, Murayama, K, Shirouzu, M, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-07-24 | | Release date: | 2007-01-24 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Crystal structure of MS0616
To be Published
|
|
