2WDR
 
 | | E. coli succinate:quinone oxidoreductase (SQR) with pentachlorophenol bound | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, FE3-S4 CLUSTER, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Ruprecht, J, Yankovskaya, V, Maklashina, E, Iwata, S, Cecchini, G. | | Deposit date: | 2009-03-25 | | Release date: | 2009-08-25 | | Last modified: | 2015-02-04 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structure of Escherichia Coli Succinate:Quinone Oxidoreductase with an Occupied and Empty Quinone- Binding Site.
J.Biol.Chem., 284, 2009
|
|
1Q4X
 
 | | Crystal Structure of Human Thyroid Hormone Receptor beta LBD in complex with specific agonist GC-24 | | Descriptor: | Thyroid hormone receptor beta-1, [4-(3-BENZYL-4-HYDROXYBENZYL)-3,5-DIMETHYLPHENOXY]ACETIC ACID | | Authors: | Borngraeber, S, Budny, M.J, Chiellini, G, Cunha-Lima, S.T, Togashi, M, Webb, P, Baxter, J.D, Scanlan, T.S, Fletterick, R.J. | | Deposit date: | 2003-08-04 | | Release date: | 2004-02-03 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Ligand selectivity by seeking hydrophobicity in thyroid hormone receptor.
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
2WDQ
 
 | | E. coli succinate:quinone oxidoreductase (SQR) with carboxin bound | | Descriptor: | 2-METHYL-N-PHENYL-5,6-DIHYDRO-1,4-OXATHIINE-3-CARBOXAMIDE, FE2/S2 (INORGANIC) CLUSTER, FE3-S4 CLUSTER, ... | | Authors: | Ruprecht, J, Yankovskaya, V, Maklashina, E, Iwata, S, Cecchini, G. | | Deposit date: | 2009-03-25 | | Release date: | 2009-08-25 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of Escherichia coli succinate:quinone oxidoreductase with an occupied and empty quinone-binding site.
J. Biol. Chem., 284, 2009
|
|
2WS3
 
 | | Crystal structure of the E. coli succinate:quinone oxidoreductase (SQR) SdhD Tyr83Phe mutant | | Descriptor: | 2-METHYL-N-PHENYL-5,6-DIHYDRO-1,4-OXATHIINE-3-CARBOXAMIDE, FE2/S2 (INORGANIC) CLUSTER, FE3-S4 CLUSTER, ... | | Authors: | Ruprecht, J, Yankovskaya, V, Maklashina, E, Iwata, S, Cecchini, G. | | Deposit date: | 2009-09-03 | | Release date: | 2010-08-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Succinate Dehydrogenase Activity
To be Published
|
|
2WU2
 
 | | Crystal structure of the E. coli succinate:quinone oxidoreductase (SQR) SdhC His84Met mutant | | Descriptor: | 2-METHYL-N-PHENYL-5,6-DIHYDRO-1,4-OXATHIINE-3-CARBOXAMIDE, FE2/S2 (INORGANIC) CLUSTER, FE3-S4 CLUSTER, ... | | Authors: | Ruprecht, J, Yankovskaya, V, Maklashina, E, Iwata, S, Cecchini, G. | | Deposit date: | 2009-09-28 | | Release date: | 2010-08-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of the E. Coli Succinate:Quinone Oxidoreductase (Sqr) Sdhc His84met Mutant
To be Published
|
|
2WQY
 
 | | Remodelling of carboxin binding to the Q-site of avian respiratory complex II | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, 2-METHYL-N-PHENYL-5,6-DIHYDRO-1,4-OXATHIINE-3-CARBOXAMIDE, AZIDE ION, ... | | Authors: | Ruprecht, J, Iwata, S, Cecchini, G. | | Deposit date: | 2009-08-27 | | Release date: | 2010-08-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Remodelling of Carboxin Binding to the Q-Site of Avian Respiratory Complex II
To be Published
|
|
1UYW
 
 | | Crystal Structure of the antiflavivirus Fab4g2 | | Descriptor: | FAB ANTIBODY HEAVY CHAIN, FAB ANTIBODY LIGHT CHAIN | | Authors: | Martinez-Fleites, C, Ortiz-Lombardia, M, Taylor, E.J, Gil-Valdes, J, Chinea, G, Davies, G. | | Deposit date: | 2004-03-03 | | Release date: | 2005-03-10 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of the Antiflavivirus Fab4G2
To be Published
|
|
1LWR
 
 | | Solution structure of the NCAM fibronectin type III module 2 | | Descriptor: | Neural Cell Adhesion Molecule 1, 140 kDa isoform | | Authors: | Kiselyov, V.V, Skladchikova, G, Hinsby, A.M, Jensen, P.H, Kulahin, N, Pedersen, N, Tsetlin, V, Poulsen, F.M, Berezin, V, Bock, E. | | Deposit date: | 2002-06-03 | | Release date: | 2003-06-03 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural basis for a direct interaction between FGFR1 and NCAM and evidence for a regulatory role of ATP
Structure, 11, 2003
|
|
4OZ6
 
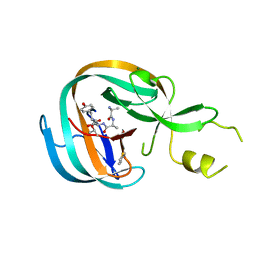 | | Structure of the Branched Intermediate in Protein Splicing | | Descriptor: | ALA-MET-ARG-TYR, MAGNESIUM ION, Mxe gyrA intein | | Authors: | Bick, M.J, Liu, Z, Frutos, S, Vila-Perello, M, Debelouchina, G.T, Darst, S.A, Muir, T.W. | | Deposit date: | 2014-02-14 | | Release date: | 2014-05-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.786 Å) | | Cite: | Structure of the branched intermediate in protein splicing.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
3P4R
 
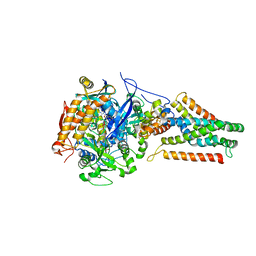 | | Crystal structure of Menaquinol:fumarate oxidoreductase in complex with glutarate | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, FE3-S4 CLUSTER, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Tomasiak, T.M, Archuleta, T.L, Andrell, J, Luna-Chavez, C, Davis, T.A, Sarwar, M, Ham, A.J, McDonald, W.H, Yankowskaya, V, Stern, H.A, Johnston, J.N, Maklashina, E, Cecchini, G, Iverson, T.M. | | Deposit date: | 2010-10-07 | | Release date: | 2010-11-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Geometric restraint drives on- and off-pathway catalysis by the Escherichia coli menaquinol:fumarate reductase.
J.Biol.Chem., 286, 2011
|
|
3P4S
 
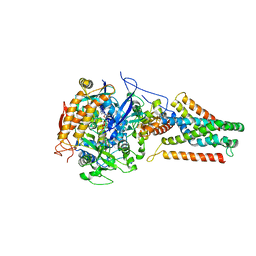 | | Crystal structure of Menaquinol:fumarate oxidoreductase in complex with a 3-nitropropionate adduct | | Descriptor: | 3-NITROPROPANOIC ACID, FE2/S2 (INORGANIC) CLUSTER, FE3-S4 CLUSTER, ... | | Authors: | Tomasiak, T.M, Archuleta, T.L, Andrell, J, Luna-Chavez, C, Davis, T.A, Sarwar, M, Ham, A.J, McDonald, W.H, Yankowskaya, V, Stern, H.A, Johnston, J.N, Maklashina, E, Cecchini, G, Iverson, T.M. | | Deposit date: | 2010-10-07 | | Release date: | 2010-12-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Geometric restraint drives on- and off-pathway catalysis by the Escherichia coli menaquinol:fumarate reductase.
J.Biol.Chem., 286, 2011
|
|
5H6N
 
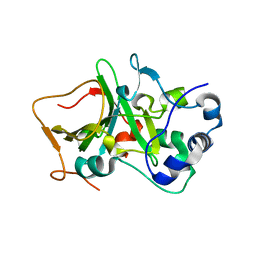 | | DNA targeting ADP-ribosyltransferase Pierisin-1, autoinhibitory form | | Descriptor: | Pierisin-1 | | Authors: | Oda, T, Hirabayashi, H, Shikauchi, G, Takamura, R, Hiraga, K, Minami, H, Hashimoto, H, Yamamoto, M, Wakabayashi, K, Sugimura, T, Shimizu, T, Sato, M. | | Deposit date: | 2016-11-14 | | Release date: | 2017-08-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis of autoinhibition and activation of the DNA-targeting ADP-ribosyltransferase pierisin-1
J. Biol. Chem., 292, 2017
|
|
4PTE
 
 | | Structure of a carvoxamide compound (15) (N-[4-(ISOQUINOLIN-7-YL)PYRIDIN-2-YL]CYCLOPROPANECARBOXAMIDE) to GSK3b | | Descriptor: | Glycogen synthase kinase-3 beta, N-[4-(isoquinolin-7-yl)pyridin-2-yl]cyclopropanecarboxamide | | Authors: | Lewis, H.A, Sivaprakasam, P, Kish, K, Pokross, M, Dubowchik, G.M. | | Deposit date: | 2014-03-10 | | Release date: | 2015-04-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.033 Å) | | Cite: | Discovery of new acylaminopyridines as GSK-3 inhibitors by a structure guided in-depth exploration of chemical space around a pyrrolopyridinone core.
Bioorg.Med.Chem.Lett., 25, 2015
|
|
4PTG
 
 | | Structure of a carboxamine compound (26) (2-{2-[(CYCLOPROPYLCARBONYL)AMINO]PYRIDIN-4-YL}-4-METHOXYPYRIMIDINE-5-CARBOXAMIDE) to GSK3b | | Descriptor: | 2-{2-[(cyclopropylcarbonyl)amino]pyridin-4-yl}-4-methoxypyrimidine-5-carboxamide, Glycogen synthase kinase-3 beta | | Authors: | Lewis, H.A, Sivaprakasam, P, Kish, K, Pokross, M, Dubowchik, G.M. | | Deposit date: | 2014-03-10 | | Release date: | 2015-04-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.361 Å) | | Cite: | Discovery of new acylaminopyridines as GSK-3 inhibitors by a structure guided in-depth exploration of chemical space around a pyrrolopyridinone core.
Bioorg.Med.Chem.Lett., 25, 2015
|
|
5F94
 
 | | Crystal structure of GSK3b in complex with Compound 15: 2-[(cyclopropylcarbonyl)amino]-N-(4-methoxypyridin-3-yl)pyridine-4-carboxamide | | Descriptor: | 2-[(cyclopropylcarbonyl)amino]-N-(4-methoxypyridin-3-yl)pyridine-4-carboxamide, Glycogen synthase kinase-3 beta | | Authors: | Lewis, H.A, Kish, K, Luo, G, Dubowchick, G.M. | | Deposit date: | 2015-12-09 | | Release date: | 2016-02-03 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Discovery of Isonicotinamides as Highly Selective, Brain Penetrable, and Orally Active Glycogen Synthase Kinase-3 Inhibitors.
J.Med.Chem., 59, 2016
|
|
3PEO
 
 | | Crystal structure of acetylcholine binding protein complexed with metocurine | | Descriptor: | 6,6',7',12'-tetramethoxy-2,2,2',2'-tetramethyltubocuraran-2,2'-diium, Soluble acetylcholine receptor | | Authors: | Talley, T.T, Harel, M, Yamauchi, G.J, Radic, Z, Hansen, S, Huxford, T, Taylor, P.W. | | Deposit date: | 2010-10-27 | | Release date: | 2011-10-12 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The curare alkaloids: analyzing the poses of complexes with the acetylcholine binding protein in relation to structure and binding energetics
To be Published
|
|
7BPN
 
 | | Solution NMR structure of NF7; de novo designed protein with a novel fold | | Descriptor: | NF7 | | Authors: | Kobayashi, N, Sugiki, T, Fujiwara, T, Minami, S, Koga, R, Chikenji, G, Koga, N. | | Deposit date: | 2020-03-23 | | Release date: | 2021-03-24 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Exploration of novel alpha-beta-protein folds through de novo design
Nat.Struct.Mol.Biol., 2023
|
|
7BPL
 
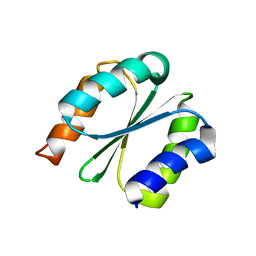 | | Solution NMR structure of NF1; de novo designed protein with a novel fold | | Descriptor: | NF1 | | Authors: | Kobayashi, N, Sugiki, T, Fujiwara, T, Minami, S, Koga, R, Chikenji, G, Koga, N. | | Deposit date: | 2020-03-23 | | Release date: | 2021-03-24 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Exploration of novel alpha-beta-protein folds through de novo design
Nat.Struct.Mol.Biol., 2023
|
|
7BPP
 
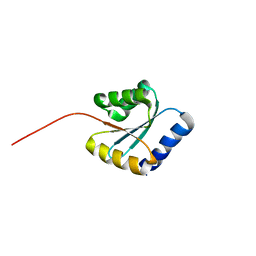 | | Solution NMR structure of NF5; de novo designed protein with a novel fold | | Descriptor: | NF5 | | Authors: | Kobayashi, N, Sugiki, T, Fujiwara, T, Minami, S, Koga, R, Chikenji, G, Koga, N. | | Deposit date: | 2020-03-23 | | Release date: | 2021-03-24 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Exploration of novel alpha-beta-protein folds through de novo design
Nat.Struct.Mol.Biol., 2023
|
|
7BPM
 
 | | Solution NMR structure of NF2; de novo designed protein with a novel fold | | Descriptor: | NF2 | | Authors: | Kobayashi, N, Sugiki, T, Fujiwara, T, Minami, S, Koga, R, Chikenji, G, Koga, N. | | Deposit date: | 2020-03-23 | | Release date: | 2021-03-24 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Exploration of novel alpha-beta-protein folds through de novo design
Nat.Struct.Mol.Biol., 2023
|
|
7BQD
 
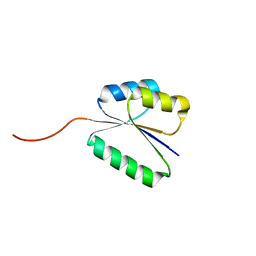 | | Solution NMR structure of NF8 (knot fold); de novo designed protein with a novel fold | | Descriptor: | NF8 | | Authors: | Kobayashi, N, Sugiki, T, Fujiwara, T, Minami, S, Koga, R, Chikenji, G, Koga, N. | | Deposit date: | 2020-03-24 | | Release date: | 2021-03-24 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Exploration of novel alpha-beta-protein folds through de novo design
Nat.Struct.Mol.Biol., 2023
|
|
5H6K
 
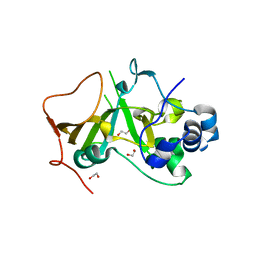 | | DNA targeting ADP-ribosyltransferase Pierisin-1 | | Descriptor: | 1,2-ETHANEDIOL, Pierisin-1 | | Authors: | Oda, T, Hirabayashi, H, Shikauchi, G, Takamura, R, Hiraga, K, Minami, H, Hashimoto, H, Yamamoto, M, Wakabayashi, K, Sugimura, T, Shimizu, T, Sato, M. | | Deposit date: | 2016-11-14 | | Release date: | 2017-08-09 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis of autoinhibition and activation of the DNA-targeting ADP-ribosyltransferase pierisin-1
J. Biol. Chem., 292, 2017
|
|
5H6L
 
 | | DNA targeting ADP-ribosyltransferase Pierisin-1 in complex with beta-NAD+ | | Descriptor: | 1,2-ETHANEDIOL, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Pierisin-1 | | Authors: | Oda, T, Hirabayashi, H, Shikauchi, G, Takamura, R, Hiraga, K, Minami, H, Hashimoto, H, Yamamoto, M, Wakabayashi, K, Sugimura, T, Shimizu, T, Sato, M. | | Deposit date: | 2016-11-14 | | Release date: | 2017-08-09 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis of autoinhibition and activation of the DNA-targeting ADP-ribosyltransferase pierisin-1
J. Biol. Chem., 292, 2017
|
|
5H6M
 
 | | DNA targeting ADP-ribosyltransferase Pierisin-1 | | Descriptor: | 1,2-ETHANEDIOL, Pierisin-1 | | Authors: | Oda, T, Hirabayashi, H, Shikauchi, G, Takamura, R, Hiraga, K, Minami, H, Hashimoto, H, Yamamoto, M, Wakabayashi, K, Sugimura, T, Shimizu, T, Sato, M. | | Deposit date: | 2016-11-14 | | Release date: | 2017-08-09 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis of autoinhibition and activation of the DNA-targeting ADP-ribosyltransferase pierisin-1
J. Biol. Chem., 292, 2017
|
|
5F95
 
 | | Crystal structure of GSK3b in complex with Compound 18: 2-[(cyclopropylcarbonyl)amino]-N-(4-phenylpyridin-3-yl)pyridine-4-carboxamide | | Descriptor: | 2-[(cyclopropylcarbonyl)amino]-N-(4-phenylpyridin-3-yl)pyridine-4-carboxamide, Glycogen synthase kinase-3 beta | | Authors: | Lewis, H.A, Kish, K, Luo, G, Dubowchick, G.M. | | Deposit date: | 2015-12-09 | | Release date: | 2016-02-03 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.525 Å) | | Cite: | Discovery of Isonicotinamides as Highly Selective, Brain Penetrable, and Orally Active Glycogen Synthase Kinase-3 Inhibitors.
J.Med.Chem., 59, 2016
|
|
