6ZEK
 
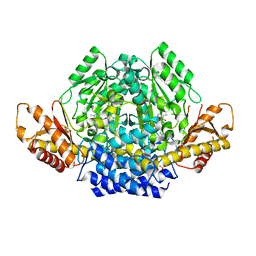 | | Crystal structure of mouse CSAD | | Descriptor: | CHLORIDE ION, COBALT (II) ION, Cysteine sulfinic acid decarboxylase, ... | | Authors: | Mahootchi, E, Raasakka, A, Haavik, J, Kursula, P. | | Deposit date: | 2020-06-16 | | Release date: | 2021-04-28 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure and substrate specificity determinants of the taurine biosynthetic enzyme cysteine sulphinic acid decarboxylase.
J.Struct.Biol., 213, 2021
|
|
1WGO
 
 | | Solution structure of the PKD domain from human VPS10 domain-containing receptor SorCS2 | | Descriptor: | VPS10 domain-containing receptor SorCS2 | | Authors: | Chikayama, E, Kigawa, T, Tochio, N, Koshiba, S, Inoue, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-05-28 | | Release date: | 2004-11-28 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the PKD domain from human VPS10 domain-containing receptor SorCS2
To be Published
|
|
7MOA
 
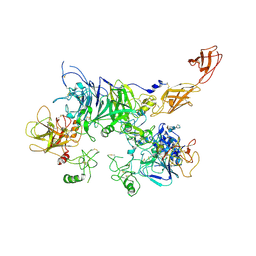 | | Cryo-EM structure of the c-MET II/HGF I complex bound with HGF II in a rigid conformation | | Descriptor: | 2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose, Hepatocyte growth factor, Hepatocyte growth factor receptor | | Authors: | Uchikawa, E, Chen, Z.M, Xiao, G.Y, Zhang, X.W, Bai, X.C. | | Deposit date: | 2021-05-01 | | Release date: | 2021-06-09 | | Last modified: | 2021-07-28 | | Method: | ELECTRON MICROSCOPY (4.9 Å) | | Cite: | Structural basis of the activation of c-MET receptor.
Nat Commun, 12, 2021
|
|
7MO7
 
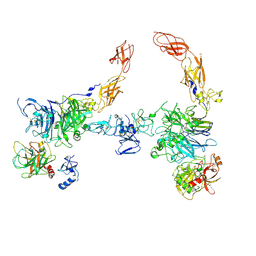 | | Cryo-EM structure of 2:2 c-MET/HGF holo-complex | | Descriptor: | 2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose, Hepatocyte growth factor, Hepatocyte growth factor receptor | | Authors: | Uchikawa, E, Chen, Z.M, Xiao, G.Y, Zhang, X.W, Bai, X.C. | | Deposit date: | 2021-05-01 | | Release date: | 2021-06-09 | | Last modified: | 2021-07-28 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Structural basis of the activation of c-MET receptor.
Nat Commun, 12, 2021
|
|
7MO9
 
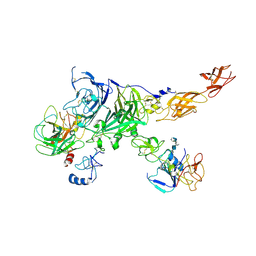 | | Cryo-EM map of the c-MET II/HGF I/HGF II (K4 and SPH) sub-complex | | Descriptor: | 2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose, Hepatocyte growth factor, Hepatocyte growth factor receptor | | Authors: | Uchikawa, E, Chen, Z.M, Xiao, G.Y, Zhang, X.W, Bai, X.C. | | Deposit date: | 2021-05-01 | | Release date: | 2021-06-09 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structural basis of the activation of c-MET receptor.
Nat Commun, 12, 2021
|
|
7MOB
 
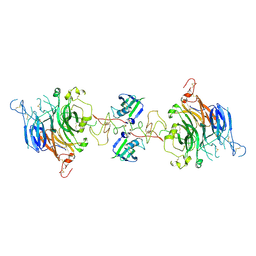 | | Cryo-EM structure of 2:2 c-MET/NK1 complex | | Descriptor: | Hepatocyte growth factor, Hepatocyte growth factor receptor | | Authors: | Uchikawa, E, Chen, Z.M, Xiao, G.Y, Zhang, X.W, Bai, X.C. | | Deposit date: | 2021-05-01 | | Release date: | 2021-06-09 | | Last modified: | 2021-07-28 | | Method: | ELECTRON MICROSCOPY (5 Å) | | Cite: | Structural basis of the activation of c-MET receptor.
Nat Commun, 12, 2021
|
|
7MO8
 
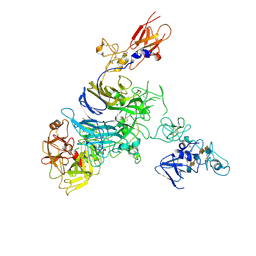 | | Cryo-EM structure of 1:1 c-MET I/HGF I complex after focused 3D refinement of holo-complex | | Descriptor: | 2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose, Hepatocyte growth factor, Hepatocyte growth factor receptor | | Authors: | Uchikawa, E, Chen, Z.M, Xiao, G.Y, Zhang, X.W, Bai, X.C. | | Deposit date: | 2021-05-01 | | Release date: | 2021-06-09 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Structural basis of the activation of c-MET receptor.
Nat Commun, 12, 2021
|
|
4WKR
 
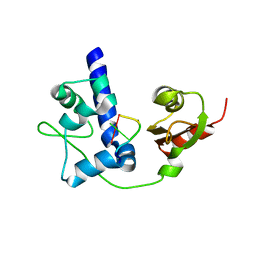 | | LaRP7 wrapping up the 3' hairpin of 7SK non-coding RNA (302-332) | | Descriptor: | 7SK GGHP4 (300-332), La-related protein 7 | | Authors: | Uchikawa, E, Natchiar, K.S, Han, X, Proux, F, Roblin, P, Zhang, E, Durand, A, Klaholz, B.P, Dock-Bregeon, A.-C. | | Deposit date: | 2014-10-03 | | Release date: | 2015-03-18 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural insight into the mechanism of stabilization of the 7SK small nuclear RNA by LARP7.
Nucleic Acids Res., 43, 2015
|
|
6PXV
 
 | | Cryo-EM structure of full-length insulin receptor bound to 4 insulin. 3D refinement was focused on the extracellular region. | | Descriptor: | Insulin, Insulin receptor | | Authors: | Uchikawa, E, Choi, E, Shang, G.J, Yu, H.T, Bai, X.C. | | Deposit date: | 2019-07-27 | | Release date: | 2019-09-04 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Activation mechanism of the insulin receptor revealed by cryo-EM structure of the fully liganded receptor-ligand complex.
Elife, 8, 2019
|
|
6PXW
 
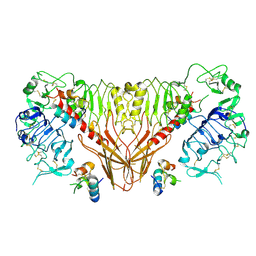 | | Cryo-EM structure of full-length insulin receptor bound to 4 insulin. 3D refinement was focused on the top part of the receptor complex. | | Descriptor: | Insulin, Insulin receptor | | Authors: | Uchikawa, E, Choi, E, Shang, G.J, Yu, H.T, Bai, X.C. | | Deposit date: | 2019-07-28 | | Release date: | 2019-09-04 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Activation mechanism of the insulin receptor revealed by cryo-EM structure of the fully liganded receptor-ligand complex.
Elife, 8, 2019
|
|
1IWQ
 
 | | Crystal Structure of MARCKS calmodulin binding domain peptide complexed with Ca2+/Calmodulin | | Descriptor: | CALCIUM ION, CALMODULIN, MARCKS | | Authors: | Yamauchi, E, Nakatsu, T, Matsubara, M, Kato, H, Taniguchi, H, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2002-05-31 | | Release date: | 2003-03-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of a MARCKS peptide containing the calmodulin-binding domain in complex with Ca(2+)-calmodulin
NAT.STRUCT.BIOL., 10, 2003
|
|
6XNN
 
 | | Crystal Structure of Mouse STING CTD complex with SR-717. | | Descriptor: | 4,5-difluoro-2-{[6-(1H-imidazol-1-yl)pyridazine-3-carbonyl]amino}benzoic acid, Stimulator of interferon genes protein | | Authors: | Chin, E.N, Yu, C, Wolan, D.W, Petrassi, H.M, Lairson, L.L. | | Deposit date: | 2020-07-03 | | Release date: | 2020-08-26 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Antitumor activity of a systemic STING-activating non-nucleotide cGAMP mimetic.
Science, 369, 2020
|
|
6XNP
 
 | | Crystal Structure of Human STING CTD complex with SR-717 | | Descriptor: | 1,2-ETHANEDIOL, 4,5-difluoro-2-{[6-(1H-imidazol-1-yl)pyridazine-3-carbonyl]amino}benzoic acid, GLYCEROL, ... | | Authors: | Chin, E.N, Yu, C, Wolan, D.W, Petrassi, H.M, Lairson, L.L. | | Deposit date: | 2020-07-03 | | Release date: | 2020-08-26 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Antitumor activity of a systemic STING-activating non-nucleotide cGAMP mimetic.
Science, 369, 2020
|
|
4YN1
 
 | | THE ATOMIC STRUCTURE OF ANOMALA CUPREA ENTOMOPOXVIRUS (ACEPV) FUSOLIN SPINDLES | | Descriptor: | 1,2-ETHANEDIOL, Fusolin, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Chiu, E, Bunker, R.D, Metcalf, P. | | Deposit date: | 2015-03-09 | | Release date: | 2015-04-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for the enhancement of virulence by viral spindles and their in vivo crystallization.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4YN2
 
 | | THE ATOMIC STRUCTURE OF WISEANA SPP ENTOMOPOXVIRUS (WSEPV) FUSOLIN SPINDLES | | Descriptor: | 1,2-ETHANEDIOL, FUSOLIN, ZINC ION, ... | | Authors: | Chiu, E, Bunker, R.D, Metcalf, P. | | Deposit date: | 2015-03-09 | | Release date: | 2015-04-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Structural basis for the enhancement of virulence by viral spindles and their in vivo crystallization.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
2ML3
 
 | |
2ML2
 
 | |
2ML1
 
 | |
7A0A
 
 | | Crystal structure of mouse CSAD in apo form | | Descriptor: | Cysteine sulfinic acid decarboxylase, SODIUM ION, SULFATE ION | | Authors: | Mahootchi, E, Raasakka, A, Haavik, J, Kursula, P. | | Deposit date: | 2020-08-07 | | Release date: | 2021-08-18 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The structure of cysteine sulphinic acid decarboxylase reveals structural determinants for substrate specificity of pyridoxal phosphate-dependent decarboxylases
To be published
|
|
3PBL
 
 | | Structure of the human dopamine D3 receptor in complex with eticlopride | | Descriptor: | 3-chloro-5-ethyl-N-{[(2S)-1-ethylpyrrolidin-2-yl]methyl}-6-hydroxy-2-methoxybenzamide, D(3) dopamine receptor, Lysozyme chimera, ... | | Authors: | Chien, E.Y.T, Liu, W, Han, G.W, Katritch, V, Zhao, Q, Cherezov, V, Stevens, R.C, Accelerated Technologies Center for Gene to 3D Structure (ATCG3D), GPCR Network (GPCR) | | Deposit date: | 2010-10-20 | | Release date: | 2010-11-03 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Structure of the human dopamine d3 receptor in complex with a d2/d3 selective antagonist.
Science, 330, 2010
|
|
1WGP
 
 | | Solution structure of the cNMP-binding domain from Arabidopsis thaliana cyclic nucleotide-regulated ion channel | | Descriptor: | Probable cyclic nucleotide-gated ion channel 6 | | Authors: | Chikayama, E, Nameki, N, Kigawa, T, Koshiba, S, Inoue, M, Tomizawa, T, Kobayashi, N, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-05-28 | | Release date: | 2004-11-28 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the cNMP-binding domain from Arabidopsis thaliana cyclic nucleotide-regulated ion channel
To be Published
|
|
1WXB
 
 | | Solution structure of the SH3 domain from human epidermal growth factor receptor pathway substrate 8-like protein | | Descriptor: | Epidermal growth factor receptor pathway substrate 8-like protein | | Authors: | Chikayama, E, Kigawa, T, Muto, Y, Koshiba, S, Inoue, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-01-20 | | Release date: | 2005-07-20 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the SH3 domain from human epidermal growth factor receptor pathway substrate 8-like protein
To be Published
|
|
1X2P
 
 | | Solution structure of the SH3 domain of the Protein arginine N-methyltransferase 2 | | Descriptor: | Protein arginine N-methyltransferase 2 | | Authors: | Chikayama, E, Kigawa, T, Saito, K, Koshiba, S, Inoue, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-04-26 | | Release date: | 2005-10-26 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the SH3 domain of the Protein arginine N-methyltransferase 2
To be Published
|
|
1WWT
 
 | | Solution structure of the TGS domain from human threonyl-tRNA synthetase | | Descriptor: | Threonyl-tRNA synthetase, cytoplasmic | | Authors: | Chikayama, E, Kigawa, T, Tomizawa, T, Koshiba, S, Inoue, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-01-18 | | Release date: | 2005-07-18 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the TGS domain from human threonyl-tRNA synthetase
To be Published
|
|
1X2Q
 
 | | Solution structure of the SH3 domain of the Signal transducing adaptor molecule 2 | | Descriptor: | Signal transducing adapter molecule 2 | | Authors: | Chikayama, E, Kigawa, T, Sato, M, Koshiba, S, Inoue, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-04-26 | | Release date: | 2005-10-26 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the SH3 domain of the Signal transducing adaptor molecule 2
To be Published
|
|
