6ORB
 
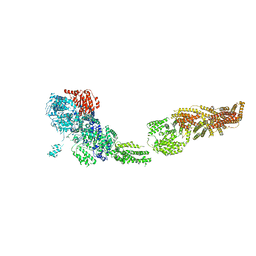 | | Full-length S. pombe Mdn1 in the presence of ATP and Rbin-1 | | Descriptor: | Midasin | | Authors: | Chen, Z, Suzuki, H, Wang, A.C, DiMaio, F, Walz, T, Kapoor, T.M. | | Deposit date: | 2019-04-29 | | Release date: | 2019-05-29 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (7.7 Å) | | Cite: | Structural Insights into Mdn1, an Essential AAA Protein Required for Ribosome Biogenesis.
Cell, 175, 2018
|
|
6OR5
 
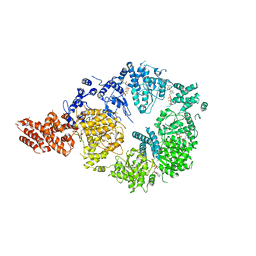 | | Full-length S. pombe Mdn1 in the presence of AMPPNP (ring region) | | Descriptor: | Midasin, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Chen, Z, Suzuki, H, Wang, A.C, DiMaio, F, Walz, T, Kapoor, T.M. | | Deposit date: | 2019-04-29 | | Release date: | 2019-05-29 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structural Insights into Mdn1, an Essential AAA Protein Required for Ribosome Biogenesis.
Cell, 175, 2018
|
|
4N78
 
 | |
7VWC
 
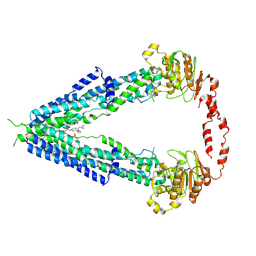 | | Cryo-EM structure of human very long-chain fatty acid ABC transporter ABCD1 | | Descriptor: | Peroxisomal Membrane Protein related,ATP-binding cassette sub-family D member 1, [(2R)-3-[2-azanylethoxy(oxidanyl)phosphoryl]oxy-2-oxidanyl-propyl] octadecanoate | | Authors: | Chen, Z.P, Xu, D, Wang, L, Mao, Y.X, Yang, L, Cheng, M.T, Hou, W.T, Chen, Y.X, Zhou, C.Z. | | Deposit date: | 2021-11-10 | | Release date: | 2022-05-18 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.53 Å) | | Cite: | Structural basis of substrate recognition and translocation by human very long-chain fatty acid transporter ABCD1.
Nat Commun, 13, 2022
|
|
7VZB
 
 | | Cryo-EM structure of C22:0-CoA bound human very long-chain fatty acid ABC transporter ABCD1 | | Descriptor: | CHOLESTEROL HEMISUCCINATE, Peroxisomal Membrane Protein related,ATP-binding cassette sub-family D member 1, S-[2-[3-[[(2R)-4-[[[(2R,3S,4R,5R)-5-(6-aminopurin-9-yl)-4-oxidanyl-3-phosphonooxy-oxolan-2-yl]methoxy-oxidanyl-phosphoryl]oxy-oxidanyl-phosphoryl]oxy-3,3-dimethyl-2-oxidanyl-butanoyl]amino]propanoylamino]ethyl] docosanethioate | | Authors: | Chen, Z.P, Xu, D, Wang, L, Mao, Y.X, Yang, L, Cheng, M.T, Hou, W.T, Chen, Y.X, Zhou, C.Z. | | Deposit date: | 2021-11-15 | | Release date: | 2022-05-18 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.59 Å) | | Cite: | Structural basis of substrate recognition and translocation by human very long-chain fatty acid transporter ABCD1.
Nat Commun, 13, 2022
|
|
7VX8
 
 | | Cryo-EM structure of ATP-bound human very long-chain fatty acid ABC transporter ABCD1 | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Peroxisomal Membrane Protein related,ATP-binding cassette sub-family D member 1 | | Authors: | Chen, Z.P, Xu, D, Wang, L, Mao, Y.X, Yang, L, Cheng, M.T, Hou, W.T, Chen, Y.X, Zhou, C.Z. | | Deposit date: | 2021-11-12 | | Release date: | 2022-05-18 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural basis of substrate recognition and translocation by human very long-chain fatty acid transporter ABCD1.
Nat Commun, 13, 2022
|
|
1HSH
 
 | |
6V08
 
 | | Crystal structure of human recombinant Beta-2 glycoprotein I (hrB2GPI) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-2-glycoprotein 1, SULFATE ION, ... | | Authors: | Chen, Z, Ruben, E.A, Planer, W, Chinnaraj, M, Zuo, X, Pengo, V, Macor, P, Tedesco, F, Pozzi, N. | | Deposit date: | 2019-11-18 | | Release date: | 2020-06-17 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | The J-elongated conformation of beta2-glycoprotein I predominates in solution: implications for our understanding of antiphospholipid syndrome.
J.Biol.Chem., 295, 2020
|
|
9JAO
 
 | | The structure of SMARCAD1 bound to the hexasome in the presence of ADP-BeFx | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, DNA (157-MER), ... | | Authors: | Chen, Z.C, Hu, P.J, Sia, Y.Y, Chen, K.J, Xia, X. | | Deposit date: | 2024-08-25 | | Release date: | 2025-04-30 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Nucleosome intermediate preference of SMARCAD1 chromatin remodeler, and the structural insights
To Be Published
|
|
4XOH
 
 | | Mechanistic insights into anchorage of the contractile ring from yeast to humans | | Descriptor: | Division mal foutue 1 protein | | Authors: | Chen, Z, Wu, J.-Q, Wang, J, Guan, R, Sun, L, Lee, I.-J, Liu, Y, Chen, M. | | Deposit date: | 2015-01-16 | | Release date: | 2015-07-15 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.801 Å) | | Cite: | Mechanistic insights into the anchorage of the contractile ring by anillin and mid1
Dev.Cell, 33, 2015
|
|
5J9Q
 
 | | Crystal structure of the NuA4 core complex | | Descriptor: | Chromatin modification-related protein EAF6, Chromatin modification-related protein YNG2, Enhancer of polycomb-like protein 1, ... | | Authors: | Chen, Z.C, Xu, P. | | Deposit date: | 2016-04-11 | | Release date: | 2016-10-26 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | The NuA4 Core Complex Acetylates Nucleosomal Histone H4 through a Double Recognition Mechanism
Mol.Cell, 63, 2016
|
|
7VOE
 
 | | Crystal structure of 5-HT2AR in complex with aripiprazole | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 5-hydroxytryptamine receptor 2A,Soluble cytochrome b562, 7-[4-[4-[2,3-bis(chloranyl)phenyl]piperazin-1-yl]butoxy]-3,4-dihydro-1H-quinolin-2-one, ... | | Authors: | Chen, Z, Fan, L, Wang, H, Yu, J, Lu, D, Qi, J, Nie, F, Luo, Z, Liu, Z, Cheng, J, Wang, S. | | Deposit date: | 2021-10-13 | | Release date: | 2021-12-22 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure-based design of a novel third-generation antipsychotic drug lead with potential antidepressant properties.
Nat.Neurosci., 25, 2022
|
|
7VOD
 
 | | Crystal structure of 5-HT2AR in complex with cariprazine | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 3-[4-[2-[4-[2,3-bis(chloranyl)phenyl]piperazin-1-yl]ethyl]cyclohexyl]-1,1-dimethyl-urea, 5-hydroxytryptamine receptor 2A,Soluble cytochrome b562, ... | | Authors: | Chen, Z, Fan, L, Wang, H, Yu, J, Lu, D, Qi, J, Nie, F, Luo, Z, Liu, Z, Cheng, J, Wang, S. | | Deposit date: | 2021-10-13 | | Release date: | 2021-12-22 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structure-based design of a novel third-generation antipsychotic drug lead with potential antidepressant properties.
Nat.Neurosci., 25, 2022
|
|
1KV9
 
 | | Structure at 1.9 A Resolution of a Quinohemoprotein Alcohol Dehydrogenase from Pseudomonas putida HK5 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ACETONE, CALCIUM ION, ... | | Authors: | Chen, Z.-W, Matsushita, K, Yamashita, T, Fujii, T, Toyama, H, Adachi, O, Bellamy, H.D, Mathews, F.S. | | Deposit date: | 2002-01-25 | | Release date: | 2002-07-10 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure at 1.9 A resolution of a quinohemoprotein alcohol dehydrogenase from Pseudomonas putida HK5.
Structure, 10, 2002
|
|
5NKP
 
 | | Crystal structure of the human KLHL3 Kelch domain in complex with a WNK3 peptide | | Descriptor: | CHLORIDE ION, CITRIC ACID, Kelch-like protein 3, ... | | Authors: | Chen, Z, Sorrell, F.J, Pinkas, D.M, Williams, E, Mathea, S, Goubin, S, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Burgess-Brown, N, Bountra, C, Bullock, A. | | Deposit date: | 2017-03-31 | | Release date: | 2017-05-03 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of the human KLHL3 Kelch domain in complex with a WNK3 peptide
To Be Published
|
|
1HSI
 
 | |
1HSG
 
 | |
8SYN
 
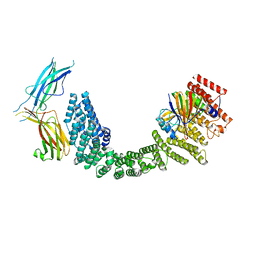 | | Human VPS35L/VPS29/VPS26C Complex | | Descriptor: | VPS35 endosomal protein-sorting factor-like, Vacuolar protein sorting-associated protein 26C, Vacuolar protein sorting-associated protein 29 | | Authors: | Chen, Z, Chen, B, Burstein, E, Han, Y. | | Deposit date: | 2023-05-25 | | Release date: | 2023-11-01 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.94 Å) | | Cite: | Structural organization of the retriever-CCC endosomal recycling complex.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8SYM
 
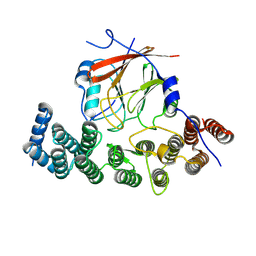 | | Human VPS29/VPS35L Complex (Locally refined map) | | Descriptor: | VPS35 endosomal protein-sorting factor-like, Vacuolar protein sorting-associated protein 29 | | Authors: | Chen, Z, Chen, B, Burstein, E, Han, Y. | | Deposit date: | 2023-05-25 | | Release date: | 2023-11-01 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural organization of the retriever-CCC endosomal recycling complex.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8SYO
 
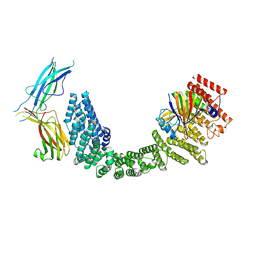 | | Human Retriever VPS35L/VPS29/VPS26C Complex (Composite Map) | | Descriptor: | VPS35 endosomal protein-sorting factor-like, Vacuolar protein sorting-associated protein 26C, Vacuolar protein sorting-associated protein 29 | | Authors: | Chen, Z, Chen, B, Burstein, E, Han, Y. | | Deposit date: | 2023-05-25 | | Release date: | 2023-11-01 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.94 Å) | | Cite: | Structural organization of the retriever-CCC endosomal recycling complex.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8D6Q
 
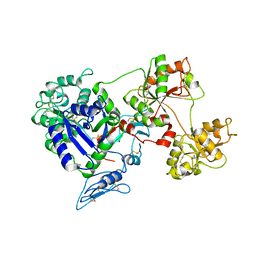 | |
8D6S
 
 | | Rana catesbeiana saxiphilin mutant - Y558A:STX (co-crystal) | | Descriptor: | Saxiphilin, [(3aS,4R,10aS)-2,6-diamino-10,10-dihydroxy-3a,4,9,10-tetrahydro-3H,8H-pyrrolo[1,2-c]purin-4-yl]methyl carbamate | | Authors: | Chen, Z, Zakrzewska, S, Minor, D.L. | | Deposit date: | 2022-06-06 | | Release date: | 2022-11-02 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Definition of a saxitoxin (STX) binding code enables discovery and characterization of the anuran saxiphilin family.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
8D6U
 
 | | Rana catesbeiana saxiphilin:F-STX (soaked) | | Descriptor: | (2P)-4-({6-[({[(3aS,4R,7R,10aS)-2,6-diamino-10,10-dihydroxy-3a,4,9,10-tetrahydro-3H,8H-pyrrolo[1,2-c]purin-4-yl]methoxy}carbonyl)amino]hexyl}carbamoyl)-2-{[4aP,9(9a)P]-6-hydroxy-3-oxo-3H-xanthen-9-yl}benzoic acid, Saxiphilin | | Authors: | Chen, Z, Zakrzewska, S, Minor, D.L. | | Deposit date: | 2022-06-06 | | Release date: | 2022-11-02 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Definition of a saxitoxin (STX) binding code enables discovery and characterization of the anuran saxiphilin family.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
8D6T
 
 | | Rana catesbeiana saxiphilin mutant - Y558I:STX (co-crystal) | | Descriptor: | CACODYLATE ION, Saxiphilin, [(3aS,4R,10aS)-2,6-diamino-10,10-dihydroxy-3a,4,9,10-tetrahydro-3H,8H-pyrrolo[1,2-c]purin-4-yl]methyl carbamate | | Authors: | Chen, Z, Zakrzewska, S, Minor, D.L. | | Deposit date: | 2022-06-06 | | Release date: | 2022-11-02 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Definition of a saxitoxin (STX) binding code enables discovery and characterization of the anuran saxiphilin family.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
8D6P
 
 | |
